Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001461A_C01 KMC001461A_c01
(674 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
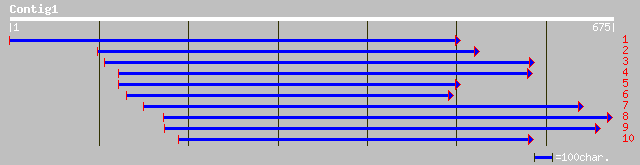
ref|NP_172679.1| hypothetical protein; protein id: At1g12150.1 [... 39 0.026
ref|NP_245379.1| unknown [Pasteurella multocida] gi|12720696|gb|... 37 0.19
pir||B38145 invariant surface glycoprotein 75 - Trypanosoma bruc... 37 0.32
gb|AAA30149.1| 75 kDa invariant surface glycoprotein 37 0.32
ref|NP_660436.1| flagellar biosynthetic protein FliR [Buchnera a... 36 0.42
>ref|NP_172679.1| hypothetical protein; protein id: At1g12150.1 [Arabidopsis
thaliana] gi|25402708|pir||E86256 hypothetical protein
[imported] - Arabidopsis thaliana
gi|10086518|gb|AAG12578.1|AC022522_11 Hypothetical
protein [Arabidopsis thaliana]
Length = 548
Score = 38.5 bits (88), Expect(2) = 0.026
Identities = 22/63 (34%), Positives = 41/63 (64%)
Frame = -2
Query: 568 RRNENKSSKAEARG*KSGVVAEEMEQKLELLLKQAEEVRAAGQKAIEEMKYCLVHKVEES 389
R+ E+ + EA + + AEE E++LEL++++ EE ++A +K EEMK ++ + +ES
Sbjct: 390 RKIESLKKETEA----AMIAAEEAEKRLELVIREVEEAKSAEEKVREEMK--MISQKQES 443
Query: 388 QFQ 380
+ Q
Sbjct: 444 KKQ 446
Score = 20.8 bits (42), Expect(2) = 0.026
Identities = 8/21 (38%), Positives = 14/21 (66%)
Frame = -1
Query: 641 HEINMEVVQLSFERERMQEKK 579
+ + ME+ L ERE +Q+K+
Sbjct: 329 NSLRMELEDLRREREELQQKE 349
>ref|NP_245379.1| unknown [Pasteurella multocida] gi|12720696|gb|AAK02526.1| unknown
[Pasteurella multocida]
Length = 229
Score = 37.4 bits (85), Expect = 0.19
Identities = 23/57 (40%), Positives = 30/57 (52%)
Frame = -2
Query: 562 NENKSSKAEARG*KSGVVAEEMEQKLELLLKQAEEVRAAGQKAIEEMKYCLVHKVEE 392
NE K+S EA VA ++E+ E + EEV+AA +EEMK KVEE
Sbjct: 50 NEVKNSAVEAAKEAKETVATKVEEVKETTAAKVEEVKAATAAKVEEMKETTAAKVEE 106
>pir||B38145 invariant surface glycoprotein 75 - Trypanosoma brucei
gi|161948|gb|AAA30148.1| 75 kDa invariant surface
glycoprotein
Length = 523
Score = 36.6 bits (83), Expect = 0.32
Identities = 23/72 (31%), Positives = 36/72 (49%)
Frame = -2
Query: 601 ERECKKRRRRTRRNENKSSKAEARG*KSGVVAEEMEQKLELLLKQAEEVRAAGQKAIEEM 422
E E K++ E + + EA + V AEE + L ++AE+ + AGQ EE
Sbjct: 307 EEEAKRQAAEKAAEEARKALEEAEARR--VAAEEQAEARRLEAEKAEKAKEAGQPVSEEK 364
Query: 421 KYCLVHKVEESQ 386
K L+ VEE++
Sbjct: 365 KKMLLEAVEEAE 376
>gb|AAA30149.1| 75 kDa invariant surface glycoprotein
Length = 325
Score = 36.6 bits (83), Expect = 0.32
Identities = 23/72 (31%), Positives = 36/72 (49%)
Frame = -2
Query: 601 ERECKKRRRRTRRNENKSSKAEARG*KSGVVAEEMEQKLELLLKQAEEVRAAGQKAIEEM 422
E E K++ E + + EA + V AEE + L ++AE+ + AGQ EE
Sbjct: 109 EEEAKRQAAEKAAEEARKALEEAEARR--VAAEEQAEARRLEAEKAEKAKEAGQPVSEEK 166
Query: 421 KYCLVHKVEESQ 386
K L+ VEE++
Sbjct: 167 KKMLLEAVEEAE 178
>ref|NP_660436.1| flagellar biosynthetic protein FliR [Buchnera aphidicola str. Sg
(Schizaphis graminum)] gi|25008465|sp|Q8KA35|FLIR_BUCAP
Flagellar biosynthetic protein fliR
gi|21622974|gb|AAM67647.1| flagellar biosynthetic
protein FliR [Buchnera aphidicola str. Sg (Schizaphis
graminum)]
Length = 258
Score = 36.2 bits (82), Expect = 0.42
Identities = 20/61 (32%), Positives = 34/61 (54%), Gaps = 6/61 (9%)
Frame = +1
Query: 436 LFALPLSLLQLVSTKVPASAPFLQPQLQIF------NLLLQLLSFYFRFFVFFFFFSCIL 597
+FALP+ + L+ST + + L PQ+ IF NLL+ +L Y+ + F FF ++
Sbjct: 185 MFALPIMIFFLISTLIMSILNRLSPQISIFSIGFPLNLLIGILILYYLMSMSFPFFKSLV 244
Query: 598 S 600
+
Sbjct: 245 N 245
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 544,469,948
Number of Sequences: 1393205
Number of extensions: 10956442
Number of successful extensions: 48291
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 40294
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46995
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29704274460
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)