Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001451A_C01 KMC001451A_c01
(566 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
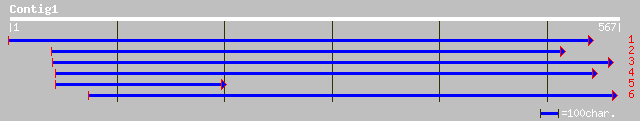
ref|NP_192851.1| putative RNA-directed RNA polymerase; protein i... 172 3e-42
pir||T30819 RNA-directed RNA polymerase (EC 2.7.7.48) - tomato g... 97 1e-19
emb|CAA09896.1| RNA-directed RNA polymerase [Petunia x hybrida] 95 7e-19
pir||T30828 RNA-directed RNA polymerase - common tobacco gi|4138... 93 2e-18
ref|NP_172932.1| RNA-dependent RNA polymerase, putative; protein... 89 4e-17
>ref|NP_192851.1| putative RNA-directed RNA polymerase; protein id: At4g11130.1
[Arabidopsis thaliana] gi|7459622|pir||T01920 probable
RNA-directed RNA polymerase (EC 2.7.7.48) - Arabidopsis
thaliana gi|3600048|gb|AAC35535.1| similar to
hypothetical proteins in Schizosaccharomyces pombe
(GB:Z98533) and C. elegans (GB:Z48334 and Z78419)
[Arabidopsis thaliana] gi|4850292|emb|CAB43048.1|
putative RNA-directed RNA polymerase [Arabidopsis
thaliana] gi|7267812|emb|CAB81214.1| putative
RNA-directed RNA polymerase [Arabidopsis thaliana]
Length = 1133
Score = 172 bits (435), Expect = 3e-42
Identities = 79/138 (57%), Positives = 107/138 (77%), Gaps = 1/138 (0%)
Frame = -1
Query: 542 AYDLNLEANGFEAFLETASSHKEMYAEKMTALMKFYGAETEDELLTGNLQNRASYLQRDN 363
AYD+ LE GFE+F+ETA +H++MY EK+T+LM +YGA E+E+LTG L+ + YL RDN
Sbjct: 982 AYDVTLEEAGFESFIETAKAHRDMYGEKLTSLMIYYGAANEEEILTGILKTKEMYLARDN 1041
Query: 362 RRYTDMKDRILISVKDLQREAKEWFEADCQ-QHEYQAMASAWYHVTYHPKYYHESSSFLS 186
RRY DMKDRI +SVKDL +EA WFE C+ + + + +ASAWY+VTY+P + E +FLS
Sbjct: 1042 RRYGDMKDRITLSVKDLHKEAMGWFEKSCEDEQQKKKLASAWYYVTYNPNHRDEKLTFLS 1101
Query: 185 FPWIVGDILLHIKSVNSK 132
FPWIVGD+LL IK+ N++
Sbjct: 1102 FPWIVGDVLLDIKAENAQ 1119
>pir||T30819 RNA-directed RNA polymerase (EC 2.7.7.48) - tomato
gi|4038592|emb|CAA71421.1| RNA-directed RNA polymerase
[Lycopersicon esculentum]
Length = 1114
Score = 97.1 bits (240), Expect = 1e-19
Identities = 50/146 (34%), Positives = 82/146 (55%), Gaps = 6/146 (4%)
Frame = -1
Query: 566 WSEKLAEDAYDLNLEANGFEAFLETASSHKEMYAEKMTALMKFYGAETEDELLTGNLQNR 387
++ +A +YD ++E +GFE +++ A +K Y K+ LM +YG +TE E+L+G +
Sbjct: 948 FTRDVARRSYDADMEVDGFEDYIDEAFDYKTEYDNKLGNLMDYYGIKTEAEILSGGIMKA 1007
Query: 386 ASYLQRDNRRYTDMKDRILISVKDLQREAKEWFEADCQQHEYQAMASAWYHVTYHPKYYH 207
+ R + I ++V+ L++EA+ WF+ + ASAWYHVTYHP Y+
Sbjct: 1008 SKTFDRRKD-----AEAISVAVRALRKEARAWFKRRNDIDDMLPKASAWYHVTYHPTYWG 1062
Query: 206 ------ESSSFLSFPWIVGDILLHIK 147
+ + F+SFPW V D L+ IK
Sbjct: 1063 CYNQGLKRAHFISFPWCVYDQLIQIK 1088
>emb|CAA09896.1| RNA-directed RNA polymerase [Petunia x hybrida]
Length = 775
Score = 94.7 bits (234), Expect = 7e-19
Identities = 52/140 (37%), Positives = 80/140 (57%), Gaps = 6/140 (4%)
Frame = -1
Query: 566 WSEKLAEDAYDLNLEANGFEAFLETASSHKEMYAEKMTALMKFYGAETEDELLTGNLQNR 387
++ +A +YD +LE +GFE +++ A +K Y +K+ LM +YG +TE E L+G +
Sbjct: 641 FTRDVARRSYDTDLEVDGFEDYIDEAFDYKSEYDKKLGNLMDYYGIKTEAETLSGGIMKA 700
Query: 386 ASYLQRDNRRYTDMKDRILISVKDLQREAKEWFEADCQQHEYQAMASAWYHVTYHPKY-- 213
+ D RR + I ++V+ L++EA+ WF+ + A ASAWYHV YHP Y
Sbjct: 701 SKTF--DRRRDA---EAIGVAVRSLRKEARTWFKRRSDIDDLLAKASAWYHVAYHPTYWG 755
Query: 212 -YHES---SSFLSFPWIVGD 165
Y+E F+SFPW V D
Sbjct: 756 CYNEGLKRDHFISFPWCVYD 775
>pir||T30828 RNA-directed RNA polymerase - common tobacco
gi|4138282|emb|CAA09697.1| RNA-directed RNA polymerase
[Nicotiana tabacum]
Length = 1116
Score = 93.2 bits (230), Expect = 2e-18
Identities = 51/151 (33%), Positives = 83/151 (54%), Gaps = 6/151 (3%)
Frame = -1
Query: 566 WSEKLAEDAYDLNLEANGFEAFLETASSHKEMYAEKMTALMKFYGAETEDELLTGNLQNR 387
++ +A +YD ++ +GFE +++ A +K Y K+ LM +YG +TE E+L+G +
Sbjct: 950 FTRDVARKSYDSDMIVDGFEDYIDEAFYYKSEYDNKLGNLMDYYGIKTEAEILSGGIMKA 1009
Query: 386 ASYLQRDNRRYTDMKDRILISVKDLQREAKEWFEADCQQHEYQAMASAWYHVTYHPKY-- 213
+ R + I ++V+ L++EA+ WF+ + A ASAWYHVTYH Y
Sbjct: 1010 SKTFDRRKD-----AEAIGVAVRCLRKEARAWFKRRSDIDDMLAKASAWYHVTYHHTYWG 1064
Query: 212 -YHES---SSFLSFPWIVGDILLHIKSVNSK 132
Y+E F+SFPW V D L+ IK ++
Sbjct: 1065 LYNEGLKRDHFISFPWCVYDQLIQIKKAKAR 1095
>ref|NP_172932.1| RNA-dependent RNA polymerase, putative; protein id: At1g14790.1
[Arabidopsis thaliana]
gi|8778232|gb|AAF79241.1|AC006917_26 F10B6.19
[Arabidopsis thaliana]
Length = 1107
Score = 89.0 bits (219), Expect = 4e-17
Identities = 54/147 (36%), Positives = 80/147 (53%), Gaps = 8/147 (5%)
Frame = -1
Query: 554 LAEDAYDLNLEANGFEAFLETASSHKEMYAEKMTALMKFYGAETEDELLTGNLQNRASYL 375
+A +YD ++E +GFE +++ A K Y K+ LM +YG +TE E+L+G +
Sbjct: 948 VASKSYDKDMEVDGFEEYVDEAFYQKANYDFKLGNLMDYYGIKTEAEILSGG-------I 1000
Query: 374 QRDNRRYTDMKDRILI--SVKDLQREAKEWFEADCQQHEYQAMASAWYHVTYHPKY---Y 210
R ++ +T +D I +V+ L++E F A ++ A ASAWYHVTYH Y Y
Sbjct: 1001 MRMSKSFTKRRDAESIGRAVRALRKETLSLFNASEEEENESAKASAWYHVTYHSSYWGLY 1060
Query: 209 HES---SSFLSFPWIVGDILLHIKSVN 138
+E FLSF W V D L+ IK N
Sbjct: 1061 NEGLNRDHFLSFAWCVYDKLVRIKKTN 1087
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 419,861,192
Number of Sequences: 1393205
Number of extensions: 8135993
Number of successful extensions: 22353
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 21861
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22314
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)