Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001446A_C01 KMC001446A_c01
(520 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
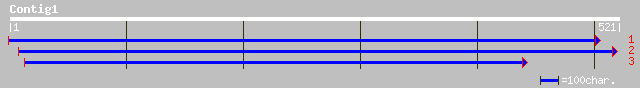
ref|NP_173662.1| hypothetical protein; protein id: At1g22460.1 [... 67 2e-10
dbj|BAB11427.1| auxin-independent growth promoter-like protein [... 55 4e-07
ref|NP_201265.2| auxin-independent growth promoter-like protein;... 55 4e-07
dbj|BAB92848.1| putative axi 1(auxin-independent growth promoter... 35 0.40
ref|NP_195730.1| putative protein; protein id: At5g01100.1 [Arab... 34 0.88
>ref|NP_173662.1| hypothetical protein; protein id: At1g22460.1 [Arabidopsis
thaliana] gi|25402965|pir||G86357 Similar to
auxin-independent growth promoter [imported] -
Arabidopsis thaliana
gi|6587842|gb|AAF18531.1|AC006551_17 Similar to
auxin-independent growth promoter [Arabidopsis thaliana]
Length = 557
Score = 66.6 bits (161), Expect = 2e-10
Identities = 29/42 (69%), Positives = 34/42 (80%)
Frame = -3
Query: 266 RLGSPRKRKGPISGTKRMDRFRSEEPFYVNPLPDCLCRTEQS 141
R GSPR+RKGP SGTK ++R RSEE FY NPLPDCLC+ + S
Sbjct: 513 RQGSPRRRKGPASGTKGLERHRSEESFYENPLPDCLCQRDPS 554
>dbj|BAB11427.1| auxin-independent growth promoter-like protein [Arabidopsis
thaliana]
Length = 539
Score = 55.5 bits (132), Expect = 4e-07
Identities = 22/41 (53%), Positives = 30/41 (72%)
Frame = -3
Query: 266 RLGSPRKRKGPISGTKRMDRFRSEEPFYVNPLPDCLCRTEQ 144
R G+PR+RKGP G K RFR+EE FY NP P+C+C +++
Sbjct: 495 RQGAPRRRKGPTQGIKGRARFRTEEAFYENPYPECICSSKE 535
>ref|NP_201265.2| auxin-independent growth promoter-like protein; protein id:
At5g64600.1, supported by cDNA: gi_18700108 [Arabidopsis
thaliana] gi|18700109|gb|AAL77666.1| AT5g64600/MUB3_12
[Arabidopsis thaliana] gi|21464583|gb|AAM52246.1|
AT5g64600/MUB3_12 [Arabidopsis thaliana]
Length = 391
Score = 55.5 bits (132), Expect = 4e-07
Identities = 22/41 (53%), Positives = 30/41 (72%)
Frame = -3
Query: 266 RLGSPRKRKGPISGTKRMDRFRSEEPFYVNPLPDCLCRTEQ 144
R G+PR+RKGP G K RFR+EE FY NP P+C+C +++
Sbjct: 347 RQGAPRRRKGPTQGIKGRARFRTEEAFYENPYPECICSSKE 387
>dbj|BAB92848.1| putative axi 1(auxin-independent growth promoter) protein [Oryza
sativa (japonica cultivar-group)]
Length = 615
Score = 35.4 bits (80), Expect = 0.40
Identities = 20/41 (48%), Positives = 23/41 (55%)
Frame = -3
Query: 266 RLGSPRKRKGPISGTKRMDRFRSEEPFYVNPLPDCLCRTEQ 144
RLG P +R +SG R EE FY NPLP CLC+ Q
Sbjct: 579 RLGGPYQR---LSGRSP----RQEEYFYANPLPGCLCKRMQ 612
>ref|NP_195730.1| putative protein; protein id: At5g01100.1 [Arabidopsis thaliana]
gi|11357840|pir||T45950 hypothetical protein F7J8.80 -
Arabidopsis thaliana gi|6759433|emb|CAB69838.1| putative
protein [Arabidopsis thaliana]
Length = 631
Score = 34.3 bits (77), Expect = 0.88
Identities = 20/47 (42%), Positives = 27/47 (56%), Gaps = 2/47 (4%)
Frame = -3
Query: 266 RLGSPRKRKGPISGTKRMDRFRSEEPFYVNPLPDCLCRT--EQSPLN 132
R+G+P R+ +G + EE FY NPLP C+C T EQ+ LN
Sbjct: 576 RIGAPYLRQPGKAGMSP----KLEENFYANPLPGCVCDTSEEQTGLN 618
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 426,895,598
Number of Sequences: 1393205
Number of extensions: 8710043
Number of successful extensions: 17983
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 17685
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17976
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16442828304
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)