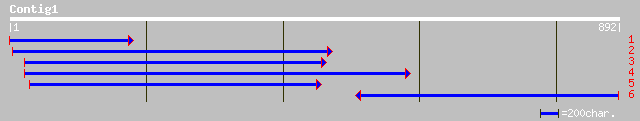
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001419A_C01 KMC001419A_c01
(889 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||C96833 hypothetical protein F18B13.24 [imported] - Arabidop... 285 8e-76
ref|NP_565231.1| expressed protein; protein id: At1g80160.1, sup... 279 4e-74
ref|NP_563973.1| expressed protein; protein id: At1g15380.1, sup... 276 4e-73
dbj|BAB62552.1| OSJNBa0089K24.9 [Oryza sativa (japonica cultivar... 216 3e-55
gb|AAK97724.1| At2g28420/T1B3.6 [Arabidopsis thaliana] gi|169744... 172 6e-42
>pir||C96833 hypothetical protein F18B13.24 [imported] - Arabidopsis thaliana
gi|5902371|gb|AAD55473.1|AC009322_13 Hypothetical
protein [Arabidopsis thaliana]
Length = 208
Score = 285 bits (728), Expect = 8e-76
Identities = 139/184 (75%), Positives = 157/184 (84%), Gaps = 2/184 (1%)
Frame = -2
Query: 837 FDHFSNFFLFV*DIFARKMKESGGNPLRLKSVNHISLICTSVEESMNFYQNVLGFIPIRR 658
+D S FL+V KMK+ GNPL +KS+NHISL+C SVEES++FYQNVLGF+PIRR
Sbjct: 24 YDRASVLFLYVSLGKKEKMKDETGNPLHIKSLNHISLLCRSVEESISFYQNVLGFLPIRR 83
Query: 657 PGSFDFDGAWLFGYGIGIHLLQAEDPENIPRKTEINPKDNHISFQCESMGAVEKKLKEME 478
P SFDFDGAWLFG+GIGIHLLQ+ +PE + +KTEINPKDNHISFQCESM AVEKKLKEME
Sbjct: 84 PDSFDFDGAWLFGHGIGIHLLQSPEPEKLLKKTEINPKDNHISFQCESMEAVEKKLKEME 143
Query: 477 IGYVRAMVEEGGIQVDQLFFHDPDGFMIEICNCDSLPVIPLAGEMVRSCSRVNLEMMQQ- 301
I YVRA+VEEGGIQVDQLFFHDPD FMIEICNCDSLPVIPLAGEM RSCSR+N+ + Q
Sbjct: 144 IEYVRAVVEEGGIQVDQLFFHDPDAFMIEICNCDSLPVIPLAGEMARSCSRLNIRQLVQP 203
Query: 300 -QIH 292
QIH
Sbjct: 204 TQIH 207
>ref|NP_565231.1| expressed protein; protein id: At1g80160.1, supported by cDNA:
21663. [Arabidopsis thaliana] gi|21554257|gb|AAM63332.1|
unknown [Arabidopsis thaliana]
Length = 167
Score = 279 bits (714), Expect = 4e-74
Identities = 133/166 (80%), Positives = 149/166 (89%), Gaps = 2/166 (1%)
Frame = -2
Query: 783 MKESGGNPLRLKSVNHISLICTSVEESMNFYQNVLGFIPIRRPGSFDFDGAWLFGYGIGI 604
MK+ GNPL +KS+NHISL+C SVEES++FYQNVLGF+PIRRP SFDFDGAWLFG+GIGI
Sbjct: 1 MKDETGNPLHIKSLNHISLLCRSVEESISFYQNVLGFLPIRRPDSFDFDGAWLFGHGIGI 60
Query: 603 HLLQAEDPENIPRKTEINPKDNHISFQCESMGAVEKKLKEMEIGYVRAMVEEGGIQVDQL 424
HLLQ+ +PE + +KTEINPKDNHISFQCESM AVEKKLKEMEI YVRA+VEEGGIQVDQL
Sbjct: 61 HLLQSPEPEKLLKKTEINPKDNHISFQCESMEAVEKKLKEMEIEYVRAVVEEGGIQVDQL 120
Query: 423 FFHDPDGFMIEICNCDSLPVIPLAGEMVRSCSRVNLEMMQQ--QIH 292
FFHDPD FMIEICNCDSLPVIPLAGEM RSCSR+N+ + Q QIH
Sbjct: 121 FFHDPDAFMIEICNCDSLPVIPLAGEMARSCSRLNIRQLVQPTQIH 166
>ref|NP_563973.1| expressed protein; protein id: At1g15380.1, supported by cDNA:
5364., supported by cDNA: gi_18252884 [Arabidopsis
thaliana] gi|25511634|pir||D86288 T24D18.8 protein -
Arabidopsis thaliana
gi|5103836|gb|AAD39666.1|AC007591_31 Is a member of the
PF|00903 gyloxalase family. ESTs gb|T44721, gb|T21844
and gb|AA395404 come from this gene. [Arabidopsis
thaliana] gi|18252885|gb|AAL62369.1| unknown protein
[Arabidopsis thaliana] gi|21387069|gb|AAM47938.1|
unknown protein [Arabidopsis thaliana]
gi|21593872|gb|AAM65839.1| unknown [Arabidopsis
thaliana]
Length = 174
Score = 276 bits (705), Expect = 4e-73
Identities = 132/172 (76%), Positives = 146/172 (84%), Gaps = 5/172 (2%)
Frame = -2
Query: 783 MKESGGNPLRLKSVNHISLICTSVEESMNFYQNVLGFIPIRRPGSFDFDGAWLFGYGIGI 604
MKE GNPL L S+NH+S++C SV+ESMNFYQ VLGFIPIRRP S +F+GAWLFG+GIGI
Sbjct: 1 MKEDAGNPLHLTSLNHVSVLCRSVDESMNFYQKVLGFIPIRRPESLNFEGAWLFGHGIGI 60
Query: 603 HLLQAEDPENIPRKTEINPKDNHISFQCESMGAVEKKLKEMEIGYVRAMVEEGGIQVDQL 424
HLL A +PE +P+KT INPKDNHISFQCESMG VEKKL+EM I YVRA+VEEGGIQVDQL
Sbjct: 61 HLLCAPEPEKLPKKTAINPKDNHISFQCESMGVVEKKLEEMGIDYVRALVEEGGIQVDQL 120
Query: 423 FFHDPDGFMIEICNCDSLPVIPLAGEMVRSCSRVNLEMM-----QQQIHQVV 283
FFHDPDGFMIEICNCDSLPV+PL GEM RSCSRV L M Q QIHQVV
Sbjct: 121 FFHDPDGFMIEICNCDSLPVVPLVGEMARSCSRVKLHQMVQPQPQTQIHQVV 172
>dbj|BAB62552.1| OSJNBa0089K24.9 [Oryza sativa (japonica cultivar-group)]
Length = 215
Score = 216 bits (551), Expect = 3e-55
Identities = 96/139 (69%), Positives = 116/139 (83%)
Frame = -2
Query: 771 GGNPLRLKSVNHISLICTSVEESMNFYQNVLGFIPIRRPGSFDFDGAWLFGYGIGIHLLQ 592
GG L L ++NH+SL+C S+ S+ FY++ LGF+ +RRPGSFDFDGAWLF YGIGIHLLQ
Sbjct: 13 GGAALPLSTLNHVSLVCRSLSTSLTFYRDFLGFVSVRRPGSFDFDGAWLFNYGIGIHLLQ 72
Query: 591 AEDPENIPRKTEINPKDNHISFQCESMGAVEKKLKEMEIGYVRAMVEEGGIQVDQLFFHD 412
AEDPE++P EINPKDNHISF CESM AV+++LKEM + YV+ VEEGG+ VDQ+FFHD
Sbjct: 73 AEDPESMPPNKEINPKDNHISFTCESMEAVQRRLKEMGVRYVQRRVEEGGVYVDQIFFHD 132
Query: 411 PDGFMIEICNCDSLPVIPL 355
PDGFMIEIC CD LPV+PL
Sbjct: 133 PDGFMIEICTCDKLPVVPL 151
>gb|AAK97724.1| At2g28420/T1B3.6 [Arabidopsis thaliana] gi|16974407|gb|AAL31129.1|
At2g28420/T1B3.6 [Arabidopsis thaliana]
Length = 184
Score = 172 bits (436), Expect = 6e-42
Identities = 75/134 (55%), Positives = 106/134 (78%), Gaps = 2/134 (1%)
Frame = -2
Query: 753 LKSVNHISLICTSVEESMNFYQNVLGFIPIRRPGSFDFDGAWLFGYGIGIHLLQAEDPEN 574
L ++NH+S +C V++S+ FY VLGF+ I RP SFDFDGAWLF YG+GIHL+QA+D +
Sbjct: 18 LMALNHVSRLCKDVKKSLKFYTKVLGFVEIERPASFDFDGAWLFNYGVGIHLVQAKDQDK 77
Query: 573 IPRKTE-INPKDNHISFQCESMGAVEKKLKEMEIGYVRAMV-EEGGIQVDQLFFHDPDGF 400
+P T+ ++P DNHISFQCE M A+EK+LKE+++ Y++ V +E +DQLFF+DPDGF
Sbjct: 78 LPSDTDHLDPMDNHISFQCEDMEALEKRLKEVKVKYIKRTVGDEKDAAIDQLFFNDPDGF 137
Query: 399 MIEICNCDSLPVIP 358
M+EICNC++L ++P
Sbjct: 138 MVEICNCENLELVP 151
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 778,236,228
Number of Sequences: 1393205
Number of extensions: 18464392
Number of successful extensions: 62653
Number of sequences better than 10.0: 195
Number of HSP's better than 10.0 without gapping: 43935
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 53255
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 48218255001
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)