Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001418A_C01 KMC001418A_c01
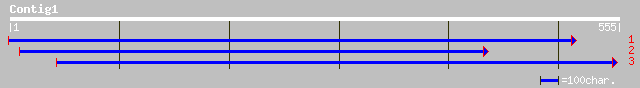
(555 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL38378.1| At2g13540/T10F5.8 [Arabidopsis thaliana] gi|23308... 238 5e-62
ref|NP_565356.1| putative cap-binding protein; protein id: At2g1... 238 5e-62
pir||C84508 probable cap-binding protein [imported] - Arabidopsi... 238 5e-62
gb|AAG54079.1| nuclear cap-binding protein CBP80 [Oryza sativa s... 191 4e-48
pdb|1H6K|A Chain A, Nuclear Cap Binding Complex gi|15988383|pdb|... 62 5e-09
>gb|AAL38378.1| At2g13540/T10F5.8 [Arabidopsis thaliana] gi|23308493|gb|AAN18216.1|
At2g13540/T10F5.8 [Arabidopsis thaliana]
Length = 848
Score = 238 bits (606), Expect = 5e-62
Identities = 110/154 (71%), Positives = 130/154 (83%)
Frame = +3
Query: 93 MSSWRSLLLRIGDKSPEYGASSDFKDHIETCFGALRREPDLSQDEILEFVLACSEQLPHK 272
MS+W++LLLRIG+K PEYG SSD+KDHIETCFG +RRE + S D++L F+L C+EQLPHK
Sbjct: 1 MSNWKTLLLRIGEKGPEYGTSSDYKDHIETCFGVIRREIERSGDQVLPFLLQCAEQLPHK 60
Query: 273 IPLYGTLIGLINLENEDLVKKFVEKTQTKFQDALDSGNCNGVRILMRLITVMMCSKVVQP 452
IPLYGTLIGL+NLENED V+K VE FQ ALDSGNCN +RIL+R +T ++CSKV+QP
Sbjct: 61 IPLYGTLIGLLNLENEDFVQKLVESVHANFQVALDSGNCNSIRILLRFMTSLLCSKVIQP 120
Query: 453 GSLVGVFETFLSSAATTVDEEKGNPLWQPCADFY 554
SL+ VFET LSSAATTVDEEKGNP WQP ADFY
Sbjct: 121 ASLIVVFETLLSSAATTVDEEKGNPSWQPQADFY 154
>ref|NP_565356.1| putative cap-binding protein; protein id: At2g13540.1, supported by
cDNA: gi_15192737, supported by cDNA: gi_17473508,
supported by cDNA: gi_8515770 [Arabidopsis thaliana]
gi|8515771|gb|AAF76167.1|AF268377_1 nuclear cap-binding
protein CBP80 [Arabidopsis thaliana]
gi|15192738|gb|AAK91588.1|AF272891_1 mRNA cap binding
protein [Arabidopsis thaliana]
gi|20198048|gb|AAD22677.2| putative cap-binding protein
[Arabidopsis thaliana]
Length = 848
Score = 238 bits (606), Expect = 5e-62
Identities = 110/154 (71%), Positives = 130/154 (83%)
Frame = +3
Query: 93 MSSWRSLLLRIGDKSPEYGASSDFKDHIETCFGALRREPDLSQDEILEFVLACSEQLPHK 272
MS+W++LLLRIG+K PEYG SSD+KDHIETCFG +RRE + S D++L F+L C+EQLPHK
Sbjct: 1 MSNWKTLLLRIGEKGPEYGTSSDYKDHIETCFGVIRREIERSGDQVLPFLLQCAEQLPHK 60
Query: 273 IPLYGTLIGLINLENEDLVKKFVEKTQTKFQDALDSGNCNGVRILMRLITVMMCSKVVQP 452
IPLYGTLIGL+NLENED V+K VE FQ ALDSGNCN +RIL+R +T ++CSKV+QP
Sbjct: 61 IPLYGTLIGLLNLENEDFVQKLVESVHANFQVALDSGNCNSIRILLRFMTSLLCSKVIQP 120
Query: 453 GSLVGVFETFLSSAATTVDEEKGNPLWQPCADFY 554
SL+ VFET LSSAATTVDEEKGNP WQP ADFY
Sbjct: 121 ASLIVVFETLLSSAATTVDEEKGNPSWQPQADFY 154
>pir||C84508 probable cap-binding protein [imported] - Arabidopsis thaliana
Length = 749
Score = 238 bits (606), Expect = 5e-62
Identities = 110/154 (71%), Positives = 130/154 (83%)
Frame = +3
Query: 93 MSSWRSLLLRIGDKSPEYGASSDFKDHIETCFGALRREPDLSQDEILEFVLACSEQLPHK 272
MS+W++LLLRIG+K PEYG SSD+KDHIETCFG +RRE + S D++L F+L C+EQLPHK
Sbjct: 1 MSNWKTLLLRIGEKGPEYGTSSDYKDHIETCFGVIRREIERSGDQVLPFLLQCAEQLPHK 60
Query: 273 IPLYGTLIGLINLENEDLVKKFVEKTQTKFQDALDSGNCNGVRILMRLITVMMCSKVVQP 452
IPLYGTLIGL+NLENED V+K VE FQ ALDSGNCN +RIL+R +T ++CSKV+QP
Sbjct: 61 IPLYGTLIGLLNLENEDFVQKLVESVHANFQVALDSGNCNSIRILLRFMTSLLCSKVIQP 120
Query: 453 GSLVGVFETFLSSAATTVDEEKGNPLWQPCADFY 554
SL+ VFET LSSAATTVDEEKGNP WQP ADFY
Sbjct: 121 ASLIVVFETLLSSAATTVDEEKGNPSWQPQADFY 154
>gb|AAG54079.1| nuclear cap-binding protein CBP80 [Oryza sativa subsp. japonica]
Length = 910
Score = 191 bits (486), Expect = 4e-48
Identities = 91/153 (59%), Positives = 113/153 (73%)
Frame = +3
Query: 96 SSWRSLLLRIGDKSPEYGASSDFKDHIETCFGALRREPDLSQDEILEFVLACSEQLPHKI 275
+ WR+LLLRIGD+ PEYG S D K+HIETC+G L RE + S+D + EF+L C++QLPHKI
Sbjct: 3 AGWRTLLLRIGDRCPEYGGSVDHKEHIETCYGVLCREYEHSKDAMFEFLLQCADQLPHKI 62
Query: 276 PLYGTLIGLINLENEDLVKKFVEKTQTKFQDALDSGNCNGVRILMRLITVMMCSKVVQPG 455
P +G LIGLINLENED K V+ T QDAL + N + +RIL+R + +MCSKVV P
Sbjct: 63 PFFGVLIGLINLENEDFSKGIVDTTHANLQDALHNENRDRIRILLRFLCGLMCSKVVLPN 122
Query: 456 SLVGVFETFLSSAATTVDEEKGNPLWQPCADFY 554
S++ FE LSSAAT +DEE GNP WQP ADFY
Sbjct: 123 SIIETFEALLSSAATILDEETGNPSWQPRADFY 155
>pdb|1H6K|A Chain A, Nuclear Cap Binding Complex gi|15988383|pdb|1H6K|B Chain
B, Nuclear Cap Binding Complex gi|15988385|pdb|1H6K|C
Chain C, Nuclear Cap Binding Complex
Length = 757
Score = 62.0 bits (149), Expect = 5e-09
Identities = 35/127 (27%), Positives = 68/127 (52%)
Frame = +3
Query: 108 SLLLRIGDKSPEYGASSDFKDHIETCFGALRREPDLSQDEILEFVLACSEQLPHKIPLYG 287
SL+ ++G+KS + + ++E G L + + +IL + + LP K+ +Y
Sbjct: 14 SLICKVGEKS-----ACSLESNLEGLAGVLEADLPNYKSKILRLLCTVARLLPEKLTIYT 68
Query: 288 TLIGLINLENEDLVKKFVEKTQTKFQDALDSGNCNGVRILMRLITVMMCSKVVQPGSLVG 467
TL+GL+N N + +FVE + +++L + N N L+R ++ ++ V+ S+V
Sbjct: 69 TLVGLLNARNYNFGGEFVEAMIRQLKESLKANNYNEAVYLVRFLSDLVNCHVIAAPSMVA 128
Query: 468 VFETFLS 488
+FE F+S
Sbjct: 129 MFENFVS 135
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 507,976,414
Number of Sequences: 1393205
Number of extensions: 10739207
Number of successful extensions: 33135
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 31840
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33114
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)