Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001416A_C01 KMC001416A_c01
(588 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
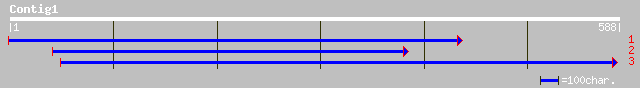
ref|XP_209680.1| similar to Tumor necrosis factor receptor super... 33 2.0
gb|AAL13562.1| GH10594p [Drosophila melanogaster] 33 2.0
ref|NP_730594.1| CG32434-PB [Drosophila melanogaster] gi|2309420... 33 2.0
ref|NP_722488.1| RIKEN cDNA 4631403P03 [Mus musculus] gi|2505451... 33 3.5
dbj|BAC31774.1| unnamed protein product [Mus musculus] 33 3.5
>ref|XP_209680.1| similar to Tumor necrosis factor receptor superfamily member 19L
precursor (Receptor expressed in lymphoid tissues) [Homo
sapiens] gi|27902291|ref|NP_776189.1| hypothetical
protein FLJ90583 [Homo sapiens]
gi|22760912|dbj|BAC11380.1| unnamed protein product
[Homo sapiens]
Length = 303
Score = 33.5 bits (75), Expect = 2.0
Identities = 25/87 (28%), Positives = 34/87 (38%)
Frame = -3
Query: 529 SVEPNLSATVSLSSHPHPEMLGAAERCYHADRSCFLPQIRLSGSRTNEQFSRTSNGSPHY 350
SV+ S SH H LG+A C H RS P +R R+ E SR G
Sbjct: 106 SVDATSSLQDGAPSHHHTVHLGSAAPCLHCSRSKRPPLVR--QGRSKEGKSRPRTGETTV 163
Query: 349 STYSRRKYGMMKMNKPLQQHHVSKPDD 269
+ R + ++ L +H P D
Sbjct: 164 FSVGRFRVTHIEKRYGLHEHRDGSPTD 190
>gb|AAL13562.1| GH10594p [Drosophila melanogaster]
Length = 1082
Score = 33.5 bits (75), Expect = 2.0
Identities = 21/66 (31%), Positives = 35/66 (52%)
Frame = -3
Query: 535 PMSVEPNLSATVSLSSHPHPEMLGAAERCYHADRSCFLPQIRLSGSRTNEQFSRTSNGSP 356
P S+ + +T +L+SHPH +L AAE Y+ ++ LPQ + + +GSP
Sbjct: 220 PASISSSTVSTSALASHPHVNLLHAAEPHYYNAQA--LPQ--------GAAYYTSYHGSP 269
Query: 355 HYSTYS 338
H +Y+
Sbjct: 270 HDLSYA 275
>ref|NP_730594.1| CG32434-PB [Drosophila melanogaster] gi|23094202|gb|AAF51675.2|
CG32434-PB [Drosophila melanogaster]
Length = 1325
Score = 33.5 bits (75), Expect = 2.0
Identities = 21/66 (31%), Positives = 35/66 (52%)
Frame = -3
Query: 535 PMSVEPNLSATVSLSSHPHPEMLGAAERCYHADRSCFLPQIRLSGSRTNEQFSRTSNGSP 356
P S+ + +T +L+SHPH +L AAE Y+ ++ LPQ + + +GSP
Sbjct: 463 PASISSSTVSTSALASHPHVNLLHAAEPHYYNAQA--LPQ--------GAAYYTSYHGSP 512
Query: 355 HYSTYS 338
H +Y+
Sbjct: 513 HDLSYA 518
>ref|NP_722488.1| RIKEN cDNA 4631403P03 [Mus musculus] gi|25054511|ref|XP_128928.2|
hypothetical protein MGC47374 [Mus musculus]
gi|23512323|gb|AAH38500.1| Unknown (protein for
MGC:47374) [Mus musculus]
Length = 294
Score = 32.7 bits (73), Expect = 3.5
Identities = 25/87 (28%), Positives = 34/87 (38%)
Frame = -3
Query: 529 SVEPNLSATVSLSSHPHPEMLGAAERCYHADRSCFLPQIRLSGSRTNEQFSRTSNGSPHY 350
SV+ S SH H LG+A C H RS P +R R+ E SR G
Sbjct: 106 SVDATSSLQDGAPSHHHTVHLGSAAPCIHCSRSKRPPLVR--QGRSKEGKSRPRPGETTV 163
Query: 349 STYSRRKYGMMKMNKPLQQHHVSKPDD 269
+ R + ++ L +H P D
Sbjct: 164 FSVGRFRVTHIEKRYGLHEHRDGSPTD 190
>dbj|BAC31774.1| unnamed protein product [Mus musculus]
Length = 303
Score = 32.7 bits (73), Expect = 3.5
Identities = 25/87 (28%), Positives = 34/87 (38%)
Frame = -3
Query: 529 SVEPNLSATVSLSSHPHPEMLGAAERCYHADRSCFLPQIRLSGSRTNEQFSRTSNGSPHY 350
SV+ S SH H LG+A C H RS P +R R+ E SR G
Sbjct: 106 SVDATSSLQDGAPSHHHTVHLGSAAPCIHCSRSKRPPLVR--QGRSKEGKSRPRPGETTV 163
Query: 349 STYSRRKYGMMKMNKPLQQHHVSKPDD 269
+ R + ++ L +H P D
Sbjct: 164 FSVGRFRVTHIEKRYGLHEHRDGSPTD 190
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 471,140,957
Number of Sequences: 1393205
Number of extensions: 9362006
Number of successful extensions: 24465
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 23557
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24446
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)