Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
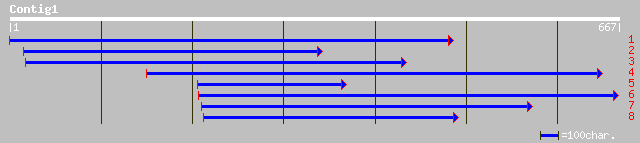
Query= KMC001413A_C01 KMC001413A_c01
(667 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM92822.1| putative signal tranduction protein [Oryza sativa... 95 1e-18
ref|NP_172965.1| unknown protein; protein id: At1g15130.1 [Arabi... 84 2e-15
gb|AAN12917.1| At1g15130/F9L1_7 [Arabidopsis thaliana] gi|290287... 84 2e-15
pir||B86285 hypothetical protein F9L1.7 [imported] - Arabidopsis... 84 2e-15
emb|CAA07235.1| extensin [Cicer arietinum] 80 2e-14
>gb|AAM92822.1| putative signal tranduction protein [Oryza sativa (japonica
cultivar-group)]
Length = 883
Score = 94.7 bits (234), Expect = 1e-18
Identities = 60/136 (44%), Positives = 67/136 (49%), Gaps = 13/136 (9%)
Frame = -2
Query: 666 PQTPYYQPDQRSDTQTNPSRPQTPYYQSAEQPPVPAYGHHPPPYGHQPP-PPYQIP---P 499
P + P + P P Y EQ P AY PPYG P PPY P P
Sbjct: 754 PPPSSHAPHAQGPYGVPPGGDSRPGYSQPEQRP--AYSQPYPPYGAPPQQPPYGAPSQQP 811
Query: 498 TSAAPYP-----PPQVHQQPPASHEYGQPAYPG-WRGPYYNAHAQQPASA---PRPPYTI 346
AP+P PP HQQPP +H+YGQ AYPG WRG YYN H QP P+PPY
Sbjct: 812 PYGAPHPGHYQQPP--HQQPP-NHDYGQQAYPGGWRGQYYNPHQPQPQPQPPYPQPPYNA 868
Query: 345 PSPYPPPHQSGYYKQQ 298
Y PPHQS YY+ Q
Sbjct: 869 QGSY-PPHQSSYYRPQ 883
Score = 44.3 bits (103), Expect = 0.001
Identities = 29/81 (35%), Positives = 36/81 (43%), Gaps = 12/81 (14%)
Frame = -2
Query: 507 IPPTSAAPYPPPQVHQQPPASHEYGQP----AYPGWRGPYYNAHAQQP-----ASAPRPP 355
+PP +P PPP P A YG P + PG+ P QP A +PP
Sbjct: 744 VPPDQNSPSPPPPSSHAPHAQGPYGVPPGGDSRPGYSQPEQRPAYSQPYPPYGAPPQQPP 803
Query: 354 YTIPSPYPP---PHQSGYYKQ 301
Y PS PP PH G+Y+Q
Sbjct: 804 YGAPSQQPPYGAPH-PGHYQQ 823
>ref|NP_172965.1| unknown protein; protein id: At1g15130.1 [Arabidopsis thaliana]
Length = 846
Score = 84.0 bits (206), Expect = 2e-15
Identities = 54/119 (45%), Positives = 60/119 (50%), Gaps = 4/119 (3%)
Frame = -2
Query: 645 PDQRSDTQTNPSRPQ--TPYYQSAEQPPVPAYGHHPPPYGHQPPPPYQIPPTSAAPYPPP 472
P +TQ NPS P PYY+ EQ P Y PPYG PPPPY P A PP
Sbjct: 747 PPPPPETQ-NPSHPHPHAPYYRPPEQMSRPGYSI--PPYG--PPPPYHTPHGQAPQPYPP 801
Query: 471 QVHQQPPASHEYGQPAYPGWR-GPYYNAHAQQPASAPRPPYTIPSPYPPPHQ-SGYYKQ 301
Q QQP +P W+ G YY+ QQP RPPY SPY PPHQ GYY+Q
Sbjct: 802 QAQQQP----------HPSWQQGSYYDPQGQQP----RPPYPGQSPYQPPHQGGGYYRQ 846
Score = 40.8 bits (94), Expect = 0.017
Identities = 32/91 (35%), Positives = 35/91 (38%), Gaps = 25/91 (27%)
Frame = -2
Query: 495 SAAPYPPPQVHQ----QPPASHEYGQPAYPGWRGPYYNAHAQQ-------PASAPRPPY- 352
S+ PYP VHQ PP E P++P PYY Q P P PPY
Sbjct: 732 SSGPYP--SVHQPTASSPPPPPETQNPSHPHPHAPYYRPPEQMSRPGYSIPPYGPPPPYH 789
Query: 351 ----TIPSPYPP-----PH----QSGYYKQQ 298
P PYPP PH Q YY Q
Sbjct: 790 TPHGQAPQPYPPQAQQQPHPSWQQGSYYDPQ 820
Score = 39.3 bits (90), Expect = 0.048
Identities = 26/81 (32%), Positives = 34/81 (41%), Gaps = 16/81 (19%)
Frame = -2
Query: 666 PQTPYYQPDQ---RSDTQTNPSRPQTPYYQ-----------SAEQPPVPAY--GHHPPPY 535
P PYY+P + R P P PY+ A+Q P P++ G + P
Sbjct: 761 PHAPYYRPPEQMSRPGYSIPPYGPPPPYHTPHGQAPQPYPPQAQQQPHPSWQQGSYYDPQ 820
Query: 534 GHQPPPPYQIPPTSAAPYPPP 472
G QP PPY +PY PP
Sbjct: 821 GQQPRPPY----PGQSPYQPP 837
>gb|AAN12917.1| At1g15130/F9L1_7 [Arabidopsis thaliana] gi|29028754|gb|AAO64756.1|
At1g15130/F9L1_7 [Arabidopsis thaliana]
Length = 846
Score = 84.0 bits (206), Expect = 2e-15
Identities = 54/119 (45%), Positives = 60/119 (50%), Gaps = 4/119 (3%)
Frame = -2
Query: 645 PDQRSDTQTNPSRPQ--TPYYQSAEQPPVPAYGHHPPPYGHQPPPPYQIPPTSAAPYPPP 472
P +TQ NPS P PYY+ EQ P Y PPYG PPPPY P A PP
Sbjct: 747 PPPPPETQ-NPSHPHPHAPYYRPPEQMSRPGYSI--PPYG--PPPPYHTPHGQAPQPYPP 801
Query: 471 QVHQQPPASHEYGQPAYPGWR-GPYYNAHAQQPASAPRPPYTIPSPYPPPHQ-SGYYKQ 301
Q QQP +P W+ G YY+ QQP RPPY SPY PPHQ GYY+Q
Sbjct: 802 QAQQQP----------HPSWQQGSYYDPQGQQP----RPPYPGQSPYQPPHQGGGYYRQ 846
Score = 40.8 bits (94), Expect = 0.017
Identities = 32/91 (35%), Positives = 35/91 (38%), Gaps = 25/91 (27%)
Frame = -2
Query: 495 SAAPYPPPQVHQ----QPPASHEYGQPAYPGWRGPYYNAHAQQ-------PASAPRPPY- 352
S+ PYP VHQ PP E P++P PYY Q P P PPY
Sbjct: 732 SSGPYP--SVHQPTASSPPPPPETQNPSHPHPHAPYYRPPEQMSRPGYSIPPYGPPPPYH 789
Query: 351 ----TIPSPYPP-----PH----QSGYYKQQ 298
P PYPP PH Q YY Q
Sbjct: 790 TPHGQAPQPYPPQAQQQPHPSWQQGSYYDPQ 820
Score = 39.3 bits (90), Expect = 0.048
Identities = 26/81 (32%), Positives = 34/81 (41%), Gaps = 16/81 (19%)
Frame = -2
Query: 666 PQTPYYQPDQ---RSDTQTNPSRPQTPYYQ-----------SAEQPPVPAY--GHHPPPY 535
P PYY+P + R P P PY+ A+Q P P++ G + P
Sbjct: 761 PHAPYYRPPEQMSRPGYSIPPYGPPPPYHTPHGQAPQPYPPQAQQQPHPSWQQGSYYDPQ 820
Query: 534 GHQPPPPYQIPPTSAAPYPPP 472
G QP PPY +PY PP
Sbjct: 821 GQQPRPPY----PGQSPYQPP 837
>pir||B86285 hypothetical protein F9L1.7 [imported] - Arabidopsis thaliana
gi|5103812|gb|AAD39642.1|AC007591_7 Similar to
gb|AJ005073 Alix (ALG-2-interacting protein X) from Mus
musculus. ESTs gb|R90133, gb|Z17944 and gb|AA605465 come
from this gene. [Arabidopsis thaliana]
Length = 816
Score = 84.0 bits (206), Expect = 2e-15
Identities = 54/119 (45%), Positives = 60/119 (50%), Gaps = 4/119 (3%)
Frame = -2
Query: 645 PDQRSDTQTNPSRPQ--TPYYQSAEQPPVPAYGHHPPPYGHQPPPPYQIPPTSAAPYPPP 472
P +TQ NPS P PYY+ EQ P Y PPYG PPPPY P A PP
Sbjct: 717 PPPPPETQ-NPSHPHPHAPYYRPPEQMSRPGYSI--PPYG--PPPPYHTPHGQAPQPYPP 771
Query: 471 QVHQQPPASHEYGQPAYPGWR-GPYYNAHAQQPASAPRPPYTIPSPYPPPHQ-SGYYKQ 301
Q QQP +P W+ G YY+ QQP RPPY SPY PPHQ GYY+Q
Sbjct: 772 QAQQQP----------HPSWQQGSYYDPQGQQP----RPPYPGQSPYQPPHQGGGYYRQ 816
Score = 40.8 bits (94), Expect = 0.017
Identities = 32/91 (35%), Positives = 35/91 (38%), Gaps = 25/91 (27%)
Frame = -2
Query: 495 SAAPYPPPQVHQ----QPPASHEYGQPAYPGWRGPYYNAHAQQ-------PASAPRPPY- 352
S+ PYP VHQ PP E P++P PYY Q P P PPY
Sbjct: 702 SSGPYP--SVHQPTASSPPPPPETQNPSHPHPHAPYYRPPEQMSRPGYSIPPYGPPPPYH 759
Query: 351 ----TIPSPYPP-----PH----QSGYYKQQ 298
P PYPP PH Q YY Q
Sbjct: 760 TPHGQAPQPYPPQAQQQPHPSWQQGSYYDPQ 790
Score = 39.3 bits (90), Expect = 0.048
Identities = 26/81 (32%), Positives = 34/81 (41%), Gaps = 16/81 (19%)
Frame = -2
Query: 666 PQTPYYQPDQ---RSDTQTNPSRPQTPYYQ-----------SAEQPPVPAY--GHHPPPY 535
P PYY+P + R P P PY+ A+Q P P++ G + P
Sbjct: 731 PHAPYYRPPEQMSRPGYSIPPYGPPPPYHTPHGQAPQPYPPQAQQQPHPSWQQGSYYDPQ 790
Query: 534 GHQPPPPYQIPPTSAAPYPPP 472
G QP PPY +PY PP
Sbjct: 791 GQQPRPPY----PGQSPYQPP 807
>emb|CAA07235.1| extensin [Cicer arietinum]
Length = 192
Score = 80.5 bits (197), Expect = 2e-14
Identities = 50/129 (38%), Positives = 58/129 (44%), Gaps = 16/129 (12%)
Frame = -2
Query: 666 PQTPYYQPDQRSDTQTNPSRPQTPYYQS----AEQPPVPAYGHHPPPYGHQPPPPYQI-- 505
P PYY +S +PS P YY+S + PP P Y H PPP PPPPY
Sbjct: 16 PPPPYYY---KSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPPPSPSPPPPYYYKS 72
Query: 504 -PPTSAAPYPPPQVHQQPPASHEYGQPAY---------PGWRGPYYNAHAQQPASAPRPP 355
PP S +P PPP ++ PP P Y P R PYY P S+P PP
Sbjct: 73 PPPPSPSP-PPPYYYKSPPPPSPSPPPPYHYQSPPPPSPTPRTPYYYKSPPPPTSSPPPP 131
Query: 354 YTIPSPYPP 328
Y SP PP
Sbjct: 132 YHYVSPPPP 140
Score = 70.1 bits (170), Expect = 3e-11
Identities = 47/126 (37%), Positives = 54/126 (42%), Gaps = 8/126 (6%)
Frame = -2
Query: 657 PYYQPDQRSDTQTNPSRPQTPYYQSAEQPPVPAYGHHPPPYGHQPPPPYQIPPTSAAPY- 481
PYY S +PS P YY+S PP P+ PP Y PPPP PP PY
Sbjct: 3 PYYY---HSPPPPSPSPPPPYYYKS---PPPPSPSPPPPYYYKSPPPPSPSPP---PPYY 53
Query: 480 ----PPPQVHQQPPASHEYGQPAYPGWRGPYYNAHAQQPASAPRPPYTIPSPYPP---PH 322
PPP PP ++ P P PYY P+ +P PPY SP PP P
Sbjct: 54 YHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYHYQSPPPPSPTPR 113
Query: 321 QSGYYK 304
YYK
Sbjct: 114 TPYYYK 119
Score = 63.5 bits (153), Expect = 2e-09
Identities = 45/120 (37%), Positives = 53/120 (43%), Gaps = 7/120 (5%)
Frame = -2
Query: 666 PQTPYYQPDQRSDTQTNPSRPQTPYYQSAEQPPVPAYGHHPPPYGH-QPPPPYQIPPTSA 490
P PYY +S +PS P PYY + PP P+ PPPY + PPPP PT
Sbjct: 64 PPPPYYY---KSPPPPSPS-PPPPYYYKSPPPPSPS---PPPPYHYQSPPPP---SPTPR 113
Query: 489 APY----PPPQVHQQPPASHEYGQPAYP--GWRGPYYNAHAQQPASAPRPPYTIPSPYPP 328
PY PPP PP H Y P P PY+ P+ +P P Y SP PP
Sbjct: 114 TPYYYKSPPPPTSSPPPPYH-YVSPPPPIKSPPPPYHYTSPPPPSPSPAPTYIYKSPPPP 172
Score = 62.0 bits (149), Expect = 7e-09
Identities = 40/99 (40%), Positives = 46/99 (46%), Gaps = 10/99 (10%)
Frame = -2
Query: 570 PVPAYGHHPPPYGHQPPPPYQI---PPTSAAPYPPPQVHQQPPASHEYGQPAYPGWRGPY 400
P P Y H PPP PPPPY PP S +P PPP ++ PP P P PY
Sbjct: 1 PPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSP-PPPYYYKSPP-------PPSPSPPPPY 52
Query: 399 YNAHAQQPASAPRPPYTI-------PSPYPPPHQSGYYK 304
Y P+ +P PPY PSP PPP+ YYK
Sbjct: 53 YYHSPPPPSPSPPPPYYYKSPPPPSPSP-PPPY---YYK 87
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 707,794,345
Number of Sequences: 1393205
Number of extensions: 21023883
Number of successful extensions: 250667
Number of sequences better than 10.0: 6611
Number of HSP's better than 10.0 without gapping: 89776
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 154478
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)