Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001384A_C01 KMC001384A_c01
(524 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
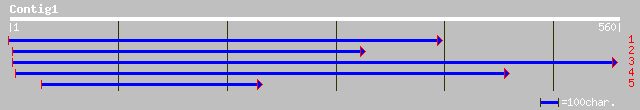
gb|AAB28462.1| extensin=nodule-specific proline-rich protein {cl... 138 4e-32
gb|AAK77904.1|AF397032_1 root nodule extensin [Pisum sativum] 129 3e-29
gb|AAK77903.1|AF397031_1 root nodule extensin [Pisum sativum] 129 3e-29
gb|AAK77905.1|AF397033_1 root nodule extensin [Pisum sativum] 127 8e-29
emb|CAA75591.1| MtN12 [Medicago truncatula] 116 2e-25
>gb|AAB28462.1| extensin=nodule-specific proline-rich protein {clone VfNDS-E}
[Vicia faba=broadbeans, root nodules, Peptide Partial,
116 aa]
Length = 116
Score = 138 bits (347), Expect = 4e-32
Identities = 57/65 (87%), Positives = 59/65 (90%), Gaps = 5/65 (7%)
Frame = -2
Query: 523 PVYHSPPPPP-PHEKPYKYASPPPPPVHTYP----HPVYHSPPPPVHSPPPPHYYYKSPP 359
PVYHSPPPPP PH+KPYKY SPPPPPVHTYP HPVYHSPPPPV+SPPPPHYYYKSPP
Sbjct: 52 PVYHSPPPPPTPHKKPYKYPSPPPPPVHTYPPHVPHPVYHSPPPPVYSPPPPHYYYKSPP 111
Query: 358 PPYHH 344
PPYHH
Sbjct: 112 PPYHH 116
Score = 82.4 bits (202), Expect = 3e-15
Identities = 39/62 (62%), Positives = 40/62 (63%), Gaps = 3/62 (4%)
Frame = -2
Query: 523 PVYHSPPPPP-PHEKPYKYASPPPPPVHTYPHP--VYHSPPPPVHSPPPPHYYYKSPPPP 353
P YHSPPPPP PH+KPYKY SPPPPPVHTYPHP VYHSPPP PP
Sbjct: 18 PFYHSPPPPPTPHKKPYKYPSPPPPPVHTYPHPHPVYHSPPP---------------PPT 62
Query: 352 YH 347
H
Sbjct: 63 PH 64
Score = 60.1 bits (144), Expect = 2e-08
Identities = 27/48 (56%), Positives = 30/48 (62%), Gaps = 3/48 (6%)
Frame = -2
Query: 478 YKYASPPPPPVHTYPHPVYHSPPPPVHSPPPPH---YYYKSPPPPYHH 344
Y Y+SPPP + +PHP YHSPPP PP PH Y Y SPPPP H
Sbjct: 2 YSYSSPPPVHTYPHPHPFYHSPPP----PPTPHKKPYKYPSPPPPPVH 45
>gb|AAK77904.1|AF397032_1 root nodule extensin [Pisum sativum]
Length = 137
Score = 129 bits (323), Expect = 3e-29
Identities = 54/65 (83%), Positives = 56/65 (86%), Gaps = 5/65 (7%)
Frame = -2
Query: 523 PVYHSPPPPP-PHEKPYKYASPPPPPVHTYPH----PVYHSPPPPVHSPPPPHYYYKSPP 359
PVYHSPPPPP PH+KPYKY SPPPPP HTYP PVYHSPPPPV+SPPPP YYYKSPP
Sbjct: 73 PVYHSPPPPPTPHKKPYKYPSPPPPPAHTYPPHVPTPVYHSPPPPVYSPPPPAYYYKSPP 132
Query: 358 PPYHH 344
PPYHH
Sbjct: 133 PPYHH 137
Score = 109 bits (273), Expect = 2e-23
Identities = 47/59 (79%), Positives = 50/59 (84%), Gaps = 2/59 (3%)
Frame = -2
Query: 523 PVYHSPPPPPPHEKPYKYASPPPPPVHTYPHP--VYHSPPPPVHSPPPPHYYYKSPPPP 353
PVYHSPPPP H+KPYKY+SPPPPPVHTYPHP VYHSPPPPVH+ P PH Y SPPPP
Sbjct: 25 PVYHSPPPP--HKKPYKYSSPPPPPVHTYPHPHPVYHSPPPPVHTYPHPHPVYHSPPPP 81
Score = 87.4 bits (215), Expect = 9e-17
Identities = 40/65 (61%), Positives = 42/65 (64%), Gaps = 16/65 (24%)
Frame = -2
Query: 493 PHEKPYKYASPPPPPVHTY--PHPVYHS--------------PPPPVHSPPPPHYYYKSP 362
PH+KPYKY SPPPPPVHTY PHPVYHS PPPPVH+ P PH Y SP
Sbjct: 2 PHKKPYKYPSPPPPPVHTYPHPHPVYHSPPPPHKKPYKYSSPPPPPVHTYPHPHPVYHSP 61
Query: 361 PPPYH 347
PPP H
Sbjct: 62 PPPVH 66
>gb|AAK77903.1|AF397031_1 root nodule extensin [Pisum sativum]
Length = 120
Score = 129 bits (323), Expect = 3e-29
Identities = 54/65 (83%), Positives = 56/65 (86%), Gaps = 5/65 (7%)
Frame = -2
Query: 523 PVYHSPPPPP-PHEKPYKYASPPPPPVHTYPH----PVYHSPPPPVHSPPPPHYYYKSPP 359
PVYHSPPPPP PH+KPYKY SPPPPP HTYP PVYHSPPPPV+SPPPP YYYKSPP
Sbjct: 56 PVYHSPPPPPTPHKKPYKYPSPPPPPAHTYPPHVPTPVYHSPPPPVYSPPPPAYYYKSPP 115
Query: 358 PPYHH 344
PPYHH
Sbjct: 116 PPYHH 120
Score = 84.7 bits (208), Expect = 6e-16
Identities = 39/63 (61%), Positives = 41/63 (64%), Gaps = 16/63 (25%)
Frame = -2
Query: 493 PHEKPYKYASPPPPPVHTY--PHPVYHS--------------PPPPVHSPPPPHYYYKSP 362
PH+KPYKY SPPPPPVHTY PHPVYHS PPPPVH+ P PH Y SP
Sbjct: 2 PHKKPYKYPSPPPPPVHTYPHPHPVYHSPPPPHKKPYKYSSPPPPPVHTYPHPHPVYHSP 61
Query: 361 PPP 353
PPP
Sbjct: 62 PPP 64
>gb|AAK77905.1|AF397033_1 root nodule extensin [Pisum sativum]
Length = 88
Score = 127 bits (319), Expect = 8e-29
Identities = 53/65 (81%), Positives = 55/65 (84%), Gaps = 5/65 (7%)
Frame = -2
Query: 523 PVYHSPPPPP-PHEKPYKYASPPPPPVHTYPH----PVYHSPPPPVHSPPPPHYYYKSPP 359
PVYHSPPPPP PH+KPYKY SPPPPP HTYP PVYHSPPPP +SPPPP YYYKSPP
Sbjct: 24 PVYHSPPPPPTPHKKPYKYPSPPPPPAHTYPPHVPTPVYHSPPPPAYSPPPPAYYYKSPP 83
Query: 358 PPYHH 344
PPYHH
Sbjct: 84 PPYHH 88
Score = 80.5 bits (197), Expect = 1e-14
Identities = 41/68 (60%), Positives = 42/68 (61%), Gaps = 21/68 (30%)
Frame = -2
Query: 493 PHEKPYKYASPPPPPVHTYPHP--VYHSPPPP---------VHSPPPP--HYY------- 374
PH+KPYKY SPPPPPVHTYPHP VYHSPPPP SPPPP H Y
Sbjct: 1 PHKKPYKYPSPPPPPVHTYPHPHPVYHSPPPPPTPHKKPYKYPSPPPPPAHTYPPHVPTP 60
Query: 373 -YKSPPPP 353
Y SPPPP
Sbjct: 61 VYHSPPPP 68
>emb|CAA75591.1| MtN12 [Medicago truncatula]
Length = 113
Score = 116 bits (290), Expect = 2e-25
Identities = 51/68 (75%), Positives = 54/68 (79%), Gaps = 8/68 (11%)
Frame = -2
Query: 523 PVYHSPPPPP----PHEKPYKYASPPPPPVHTYP----HPVYHSPPPPVHSPPPPHYYYK 368
PVYHSPPPP PH KP ++ PPPPVHTYP HPVYHSPPPPVHSPPPPHYYYK
Sbjct: 48 PVYHSPPPPVHTYVPHPKPVYHS--PPPPVHTYPPHVPHPVYHSPPPPVHSPPPPHYYYK 105
Query: 367 SPPPPYHH 344
SPPPPYH+
Sbjct: 106 SPPPPYHN 113
Score = 73.2 bits (178), Expect = 2e-12
Identities = 30/42 (71%), Positives = 31/42 (73%)
Frame = -2
Query: 472 YASPPPPPVHTYPHPVYHSPPPPVHSPPPPHYYYKSPPPPYH 347
Y SPPPP HTYP PVYHSPPPPVH+ P P Y SPPPP H
Sbjct: 2 YHSPPPPVHHTYPKPVYHSPPPPVHTYPHPKPVYHSPPPPVH 43
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 578,432,872
Number of Sequences: 1393205
Number of extensions: 18481398
Number of successful extensions: 311654
Number of sequences better than 10.0: 5479
Number of HSP's better than 10.0 without gapping: 85528
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 191224
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17019769648
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)