Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001374A_C01 KMC001374A_c01
(567 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
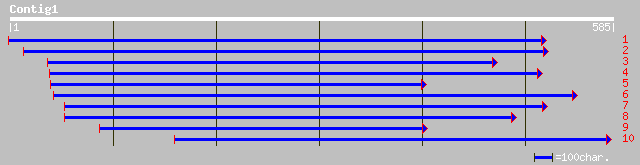
ref|NP_177383.1| unknown protein; protein id: At1g72390.1 [Arabi... 90 2e-17
gb|AAK52107.1|AC079936_3 Conserved hypothetical protein [Oryza s... 54 1e-06
ref|NP_477201.1| Deformed CG2189-PA gi|4389426|gb|AAD19796.1| ho... 39 0.045
sp|P07548|HMDF_DROME Homeotic deformed protein gi|84925|pir||A26... 37 0.17
prf||1305251A Deformed gene 37 0.17
>ref|NP_177383.1| unknown protein; protein id: At1g72390.1 [Arabidopsis thaliana]
gi|25406173|pir||H96747 unknown protein T10D10.14
[imported] - Arabidopsis thaliana
gi|12325276|gb|AAG52583.1|AC016529_14 unknown protein;
66073-62011 [Arabidopsis thaliana]
Length = 1088
Score = 89.7 bits (221), Expect = 2e-17
Identities = 48/74 (64%), Positives = 56/74 (74%), Gaps = 10/74 (13%)
Frame = -1
Query: 567 PQQMSQRTPMSPQQM----------SSGAIHAMSTGNPEACPASPQLSSQTLGSVSSMAN 418
PQQMSQRTPMSPQQ+ SSGA+H MST N E CPASPQLSSQT+GSV S+ N
Sbjct: 1012 PQQMSQRTPMSPQQVNQRTPMSPQISSGAMHPMSTSNLEGCPASPQLSSQTMGSVGSITN 1071
Query: 417 SPMDMQGVNKSNNA 376
SPM++QG K+N+A
Sbjct: 1072 SPMELQG-PKNNSA 1084
>gb|AAK52107.1|AC079936_3 Conserved hypothetical protein [Oryza sativa]
gi|15451622|gb|AAK98746.1|AC091734_3 Hypothetical protein
[Oryza sativa]
Length = 1272
Score = 53.9 bits (128), Expect = 1e-06
Identities = 32/65 (49%), Positives = 45/65 (69%), Gaps = 5/65 (7%)
Frame = -1
Query: 567 PQQMSQRTPM----SPQQMSSGAIHAMSTGNPEACPASPQLSSQTLGSVSSMANSPMD-M 403
PQQM+ +P + QQ+++ I+ ++T P P SPQLSSQT GSV+S+ANSPM+ +
Sbjct: 1205 PQQMAAMSPQLSSGTMQQVNNNVINHVATPGP---PPSPQLSSQTHGSVNSIANSPMEQL 1261
Query: 402 QGVNK 388
QG NK
Sbjct: 1262 QGANK 1266
>ref|NP_477201.1| Deformed CG2189-PA gi|4389426|gb|AAD19796.1| homeodomain protein
[Drosophila melanogaster] gi|23170614|gb|AAF54083.2|
CG2189-PA [Drosophila melanogaster]
Length = 586
Score = 38.9 bits (89), Expect = 0.045
Identities = 21/67 (31%), Positives = 38/67 (56%), Gaps = 5/67 (7%)
Frame = -1
Query: 564 QQMSQRTPMSPQQMSSGAIHAM-----STGNPEACPASPQLSSQTLGSVSSMANSPMDMQ 400
QQ +Q+TP+ + + S ++ ++ S GNP PA+P+ +S GS ++N+ +
Sbjct: 471 QQQTQQTPVMNECIRSDSLESIGDVSSSLGNPPYIPAAPETTSSYPGSQQHLSNNNNNGS 530
Query: 399 GVNKSNN 379
G N +NN
Sbjct: 531 GNNNNNN 537
>sp|P07548|HMDF_DROME Homeotic deformed protein gi|84925|pir||A26638 homeotic protein Dfd
- fruit fly (Drosophila melanogaster)
gi|7837|emb|CAA28782.1| Dfd reading frame (AA 1-590)
[Drosophila melanogaster]
Length = 590
Score = 37.0 bits (84), Expect = 0.17
Identities = 20/66 (30%), Positives = 37/66 (55%), Gaps = 5/66 (7%)
Frame = -1
Query: 561 QMSQRTPMSPQQMSSGAIHAM-----STGNPEACPASPQLSSQTLGSVSSMANSPMDMQG 397
Q +Q+TP+ + + S ++ ++ S GNP PA+P+ +S GS ++N+ + G
Sbjct: 475 QQTQQTPVMNECIRSDSLESIGDVSSSLGNPPYIPAAPETTSSYPGSQQHLSNNNNNGSG 534
Query: 396 VNKSNN 379
N +NN
Sbjct: 535 NNNNNN 540
>prf||1305251A Deformed gene
Length = 590
Score = 37.0 bits (84), Expect = 0.17
Identities = 20/66 (30%), Positives = 37/66 (55%), Gaps = 5/66 (7%)
Frame = -1
Query: 561 QMSQRTPMSPQQMSSGAIHAM-----STGNPEACPASPQLSSQTLGSVSSMANSPMDMQG 397
Q +Q+TP+ + + S ++ ++ S GNP PA+P+ +S GS ++N+ + G
Sbjct: 475 QQTQQTPVMNECIRSDSLESIGDVSSSLGNPPYIPAAPETTSSYPGSQQHLSNNNNNGSG 534
Query: 396 VNKSNN 379
N +NN
Sbjct: 535 NNNNNN 540
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 456,987,906
Number of Sequences: 1393205
Number of extensions: 9316218
Number of successful extensions: 33060
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 31269
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33029
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)