Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001371A_C01 KMC001371A_c01
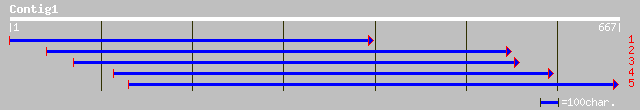
(665 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568968.1| topoisomerase, putative; protein id: At5g63190.... 171 8e-42
gb|AAM63106.1| topoisomerase-like protein [Arabidopsis thaliana] 171 8e-42
dbj|BAB10561.1| topoisomerase-like protein [Arabidopsis thaliana] 171 8e-42
ref|NP_190411.1| putative protein; protein id: At3g48390.1, supp... 167 9e-41
emb|CAD41105.1| OSJNBb0011N17.22 [Oryza sativa (japonica cultiva... 162 4e-39
>ref|NP_568968.1| topoisomerase, putative; protein id: At5g63190.1, supported by
cDNA: 19433. [Arabidopsis thaliana]
gi|14532646|gb|AAK64051.1| putative topoisomerase
[Arabidopsis thaliana] gi|23296935|gb|AAN13205.1|
putative topoisomerase [Arabidopsis thaliana]
Length = 702
Score = 171 bits (433), Expect = 8e-42
Identities = 82/109 (75%), Positives = 89/109 (81%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDRMLDLLQECFSEGLITIN 485
EEYE+ G SEACQCIRDLGM FFNHEVVKKALVMAMEKQNDR+L+LL+ECF EGLIT N
Sbjct: 593 EEYETGGVTSEACQCIRDLGMPFFNHEVVKKALVMAMEKQNDRLLNLLEECFGEGLITTN 652
Query: 484 QMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSFDSSTT 338
QMT+GF RV DSLDDL+LDIPNAKEKF Y A GW+LP F S T
Sbjct: 653 QMTKGFGRVNDSLDDLSLDIPNAKEKFELYASHAMDNGWILPEFGISAT 701
Score = 109 bits (272), Expect = 4e-23
Identities = 58/106 (54%), Positives = 77/106 (71%), Gaps = 2/106 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
+EY +G EAC+CIR+LG+SFF+HEVVK+ALV+AM+ +L LL+E EGLI+
Sbjct: 296 KEYVENGDTYEACRCIRELGVSFFHHEVVKRALVLAMDSPTAESLVLKLLKETAEEGLIS 355
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
+QM +GF RV +SLDDLALDIP+AK+ F V +A + GWL SF
Sbjct: 356 SSQMVKGFFRVAESLDDLALDIPSAKKLFDSIVPKAISGGWLDDSF 401
Score = 50.8 bits (120), Expect = 2e-05
Identities = 33/106 (31%), Positives = 55/106 (51%), Gaps = 2/106 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
+EY S+G V A +R+LG S ++ K+ + MAM++ + M +L +I
Sbjct: 132 DEYFSTGDVKVAASDLRELGSSEYHPYFTKRLVSMAMDRHDKEKEMASVLLSALYADVIL 191
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
+Q+ GF R+ S+DDLA+DI +A ++ +A L P F
Sbjct: 192 PDQIRDGFIRLLRSVDDLAVDILDAVNVLALFIARAIVDEILPPVF 237
Score = 44.7 bits (104), Expect = 0.001
Identities = 26/104 (25%), Positives = 50/104 (48%), Gaps = 2/104 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQN--DRMLDLLQECFSEGLIT 491
+EY S + E + ++DLG +N +K+ + +A++++N M +L L +
Sbjct: 430 QEYFLSDDIPELIRSLQDLGAPEYNPVFLKRLITLALDRKNREKEMASVLLSALHMELFS 489
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLP 359
GF + +S +D ALDI +A + ++ +A L P
Sbjct: 490 TEDFINGFIMLLESAEDTALDIMDASNELALFLARAVIDDVLAP 533
>gb|AAM63106.1| topoisomerase-like protein [Arabidopsis thaliana]
Length = 702
Score = 171 bits (433), Expect = 8e-42
Identities = 82/109 (75%), Positives = 89/109 (81%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDRMLDLLQECFSEGLITIN 485
EEYE+ G SEACQCIRDLGM FFNHEVVKKALVMAMEKQNDR+L+LL+ECF EGLIT N
Sbjct: 593 EEYETGGVTSEACQCIRDLGMPFFNHEVVKKALVMAMEKQNDRLLNLLEECFGEGLITTN 652
Query: 484 QMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSFDSSTT 338
QMT+GF RV DSLDDL+LDIPNAKEKF Y A GW+LP F S T
Sbjct: 653 QMTKGFGRVNDSLDDLSLDIPNAKEKFELYASHAMDNGWILPEFGISAT 701
Score = 109 bits (272), Expect = 4e-23
Identities = 58/106 (54%), Positives = 77/106 (71%), Gaps = 2/106 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
+EY +G EAC+CIR+LG+SFF+HEVVK+ALV+AM+ +L LL+E EGLI+
Sbjct: 296 KEYVENGDTYEACRCIRELGVSFFHHEVVKRALVLAMDSPTAESLVLKLLKETAEEGLIS 355
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
+QM +GF RV +SLDDLALDIP+AK+ F V +A + GWL SF
Sbjct: 356 SSQMVKGFFRVAESLDDLALDIPSAKKLFDSIVPKAISGGWLDDSF 401
Score = 50.8 bits (120), Expect = 2e-05
Identities = 33/106 (31%), Positives = 55/106 (51%), Gaps = 2/106 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
+EY S+G V A +R+LG S ++ K+ + MAM++ + M +L +I
Sbjct: 132 DEYFSTGDVKVAASDLRELGSSEYHPYFTKRLVSMAMDRHDKEKEMASVLLSALYADVIL 191
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
+Q+ GF R+ S+DDLA+DI +A ++ +A L P F
Sbjct: 192 PDQIRDGFIRLLRSVDDLAVDILDAVNVLALFIARAIVDEILPPVF 237
Score = 42.4 bits (98), Expect = 0.006
Identities = 25/104 (24%), Positives = 49/104 (47%), Gaps = 2/104 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQ--NDRMLDLLQECFSEGLIT 491
+EY S + E + ++DLG +N +K+ + +A++++ M +L L +
Sbjct: 430 QEYFLSDDIPELIRSLQDLGAPEYNPVFLKRLITLALDRKXREKEMASVLLSALHMELFS 489
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLP 359
GF + +S +D ALDI +A + ++ +A L P
Sbjct: 490 TEDFINGFIMLLESAEDTALDIMDASNELALFLARAVIDDVLAP 533
>dbj|BAB10561.1| topoisomerase-like protein [Arabidopsis thaliana]
Length = 729
Score = 171 bits (433), Expect = 8e-42
Identities = 82/109 (75%), Positives = 89/109 (81%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDRMLDLLQECFSEGLITIN 485
EEYE+ G SEACQCIRDLGM FFNHEVVKKALVMAMEKQNDR+L+LL+ECF EGLIT N
Sbjct: 620 EEYETGGVTSEACQCIRDLGMPFFNHEVVKKALVMAMEKQNDRLLNLLEECFGEGLITTN 679
Query: 484 QMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSFDSSTT 338
QMT+GF RV DSLDDL+LDIPNAKEKF Y A GW+LP F S T
Sbjct: 680 QMTKGFGRVNDSLDDLSLDIPNAKEKFELYASHAMDNGWILPEFGISAT 728
Score = 109 bits (272), Expect = 4e-23
Identities = 58/106 (54%), Positives = 77/106 (71%), Gaps = 2/106 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
+EY +G EAC+CIR+LG+SFF+HEVVK+ALV+AM+ +L LL+E EGLI+
Sbjct: 323 KEYVENGDTYEACRCIRELGVSFFHHEVVKRALVLAMDSPTAESLVLKLLKETAEEGLIS 382
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
+QM +GF RV +SLDDLALDIP+AK+ F V +A + GWL SF
Sbjct: 383 SSQMVKGFFRVAESLDDLALDIPSAKKLFDSIVPKAISGGWLDDSF 428
Score = 50.8 bits (120), Expect = 2e-05
Identities = 33/106 (31%), Positives = 55/106 (51%), Gaps = 2/106 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
+EY S+G V A +R+LG S ++ K+ + MAM++ + M +L +I
Sbjct: 159 DEYFSTGDVKVAASDLRELGSSEYHPYFTKRLVSMAMDRHDKEKEMASVLLSALYADVIL 218
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
+Q+ GF R+ S+DDLA+DI +A ++ +A L P F
Sbjct: 219 PDQIRDGFIRLLRSVDDLAVDILDAVNVLALFIARAIVDEILPPVF 264
Score = 44.7 bits (104), Expect = 0.001
Identities = 26/104 (25%), Positives = 50/104 (48%), Gaps = 2/104 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQN--DRMLDLLQECFSEGLIT 491
+EY S + E + ++DLG +N +K+ + +A++++N M +L L +
Sbjct: 457 QEYFLSDDIPELIRSLQDLGAPEYNPVFLKRLITLALDRKNREKEMASVLLSALHMELFS 516
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLP 359
GF + +S +D ALDI +A + ++ +A L P
Sbjct: 517 TEDFINGFIMLLESAEDTALDIMDASNELALFLARAVIDDVLAP 560
>ref|NP_190411.1| putative protein; protein id: At3g48390.1, supported by cDNA:
gi_18377840 [Arabidopsis thaliana]
gi|7487539|pir||T06703 hypothetical protein T29H11.90 -
Arabidopsis thaliana gi|4678349|emb|CAB41159.1| putative
protein [Arabidopsis thaliana]
gi|18377841|gb|AAL67107.1| AT3g48390/T29H11_90
[Arabidopsis thaliana] gi|25090070|gb|AAN72220.1|
At3g48390/T29H11_90 [Arabidopsis thaliana]
Length = 633
Score = 167 bits (424), Expect = 9e-41
Identities = 81/108 (75%), Positives = 91/108 (84%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDRMLDLLQECFSEGLITIN 485
EEYE G +SEAC+CIRDLGM FFNHEVVKKALVMAMEK+NDRML+LLQECF+EG+IT N
Sbjct: 524 EEYEVGGVISEACRCIRDLGMPFFNHEVVKKALVMAMEKKNDRMLNLLQECFAEGIITTN 583
Query: 484 QMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSFDSST 341
QMT+GF RVKDSLDDL+LDIPNA+EKF YV A+ GWL F ST
Sbjct: 584 QMTKGFGRVKDSLDDLSLDIPNAEEKFNSYVAHAEENGWLHRDFGCST 631
Score = 105 bits (263), Expect = 4e-22
Identities = 54/101 (53%), Positives = 71/101 (69%), Gaps = 2/101 (1%)
Frame = -2
Query: 661 EYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLITI 488
EY +G EAC+CIR+LG+SFF+HE+VK LV+ ME + +L LL+E EGLI+
Sbjct: 231 EYVENGDTREACRCIRELGVSFFHHEIVKSGLVLVMESRTSEPLILKLLKEATEEGLISS 290
Query: 487 NQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWL 365
+QM +GF+RV DSLDDL+LDIP+AK F V +A GWL
Sbjct: 291 SQMAKGFSRVADSLDDLSLDIPSAKTLFESIVPKAIIGGWL 331
Score = 55.5 bits (132), Expect = 6e-07
Identities = 36/107 (33%), Positives = 61/107 (56%), Gaps = 3/107 (2%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAME---KQNDRMLDLLQECFSEGLI 494
+EY SSG V A + DLG+S ++ VK+ + MAM+ K+ ++ LL ++ ++
Sbjct: 66 DEYFSSGDVEVAASDLMDLGLSEYHPYFVKRLVSMAMDRGNKEKEKASVLLSRLYAL-VV 124
Query: 493 TINQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
+ +Q+ GF R+ +S+ DLALDIP+A ++ +A L P F
Sbjct: 125 SPDQIRVGFIRLLESVGDLALDIPDAVNVLALFIARAIVDEILPPVF 171
Score = 43.9 bits (102), Expect = 0.002
Identities = 26/104 (25%), Positives = 49/104 (47%), Gaps = 2/104 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQN--DRMLDLLQECFSEGLIT 491
+EY S + E + + DLG+ +N +KK + +AM+++N M + + +
Sbjct: 361 QEYFLSDDIPELIRSLEDLGLPEYNPVFLKKLITLAMDRKNKEKEMASVFLASLHMEMFS 420
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLP 359
GF + +S +D ALDI A ++ ++ +A L P
Sbjct: 421 TEDFINGFIMLLESAEDTALDILAASDELALFLARAVIDDVLAP 464
>emb|CAD41105.1| OSJNBb0011N17.22 [Oryza sativa (japonica cultivar-group)]
Length = 662
Score = 162 bits (410), Expect = 4e-39
Identities = 80/105 (76%), Positives = 89/105 (84%), Gaps = 1/105 (0%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQND-RMLDLLQECFSEGLITI 488
EEY + G + EACQCIRDLGM FFNHEVVKKALVMAMEK+N+ R+L LLQECF EGLITI
Sbjct: 557 EEYNTGGDLGEACQCIRDLGMPFFNHEVVKKALVMAMEKENEARILALLQECFGEGLITI 616
Query: 487 NQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
NQMT GFTRVK+ LDDL LDIPNA+EKFG YV+ A +GWLLP F
Sbjct: 617 NQMTLGFTRVKEGLDDLILDIPNAQEKFGAYVDLATERGWLLPPF 661
Score = 100 bits (248), Expect = 2e-20
Identities = 53/110 (48%), Positives = 79/110 (71%), Gaps = 2/110 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
+EY SG + EA +CIR+LG+ FF+HEVVK+AL ++ME + + +L LL+E + LI+
Sbjct: 259 KEYIESGDIDEAFRCIRELGLPFFHHEVVKRALTLSMENLSSQPLILKLLKESTAGCLIS 318
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSFDSST 341
NQM++GF R+ +S+DDL+LDIP+AK F V A ++GWL SF +S+
Sbjct: 319 SNQMSKGFCRLAESIDDLSLDIPSAKILFDKLVLTATSEGWLDASFTTSS 368
Score = 55.8 bits (133), Expect = 5e-07
Identities = 34/106 (32%), Positives = 57/106 (53%), Gaps = 2/106 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQNDR--MLDLLQECFSEGLIT 491
EEY S+G V A +R LG F+ +KK + MAM++ + M +L L+
Sbjct: 95 EEYFSTGDVELAASELRSLGSDQFHSYFIKKLISMAMDRHDKEKEMASILLSALYADLLG 154
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLPSF 353
++M++GF + +S +DL++DIP+A + +V +A L P F
Sbjct: 155 SSKMSEGFMMLLESTEDLSVDIPDAIDVLSVFVARAVVDEILPPVF 200
Score = 42.4 bits (98), Expect = 0.006
Identities = 28/104 (26%), Positives = 51/104 (48%), Gaps = 2/104 (1%)
Frame = -2
Query: 664 EEYESSGFVSEACQCIRDLGMSFFNHEVVKKALVMAMEKQN--DRMLDLLQECFSEGLIT 491
+EY S V E +++L +N +KK + +AM+++N M L S L +
Sbjct: 394 QEYFLSDDVPELIISLQELSAPEYNPIFLKKLITLAMDRKNREKEMASALLSSLSLELFS 453
Query: 490 INQMTQGFTRVKDSLDDLALDIPNAKEKFGFYVEQAQAKGWLLP 359
+ + +GF + S +D ALDI +A + ++ +A L+P
Sbjct: 454 TDDIMKGFILLLQSAEDTALDIVDAPSELALFLARAVIDEVLIP 497
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 568,638,412
Number of Sequences: 1393205
Number of extensions: 12358013
Number of successful extensions: 27203
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 26296
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27128
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)