Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001344A_C03 KMC001344A_c03
(683 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
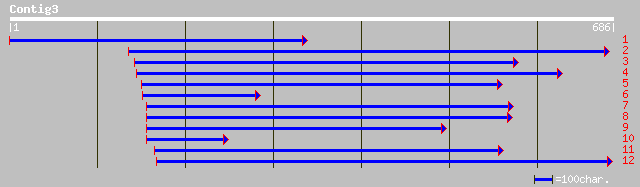
dbj|BAB21187.1| contains ESTs D22419(C10991),AU100646(C10991)~si... 95 1e-18
ref|NP_029947.2| E2, ubiquitin-conjugating enzyme, putative; pro... 94 2e-18
ref|NP_179284.1| putative ubiquitin-conjugating enzyme; protein ... 87 2e-16
dbj|BAB90019.1| putative ubiquitin-conjugating protein [Oryza sa... 74 2e-12
dbj|BAA96583.1| unnamed protein product [Oryza sativa (japonica ... 58 1e-07
>dbj|BAB21187.1| contains ESTs D22419(C10991),AU100646(C10991)~similar to Arabidopsis
thaliana chromosome 2, At2g16920~unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|14090383|dbj|BAB55541.1| P0037C04.30 [Oryza sativa
(japonica cultivar-group)]
Length = 1067
Score = 94.7 bits (234), Expect = 1e-18
Identities = 44/92 (47%), Positives = 58/92 (62%)
Frame = -3
Query: 681 MLYLLRSPPMHFEALVEEHFRQRSKQILLACEAYLEGAPIGYAFDDDGKTEHGNLKGAST 502
MLY+LR PPMHFE + HF +R K IL ACEAYL+G +G DD TE + S
Sbjct: 969 MLYILRRPPMHFEDFAKSHFSKRGKYILKACEAYLQGNGVGTLTDDACTTERSKEQPCSV 1028
Query: 501 GFKIMLAKLLPKLVEAFSDKGIDCHQFAEMQK 406
GFK+ LAK++P+L+ A D G +C Q+ + K
Sbjct: 1029 GFKLALAKIMPRLITALKDAGANCDQYEHLGK 1060
>ref|NP_029947.2| E2, ubiquitin-conjugating enzyme, putative; protein id: At2g33770.1,
supported by cDNA: gi_18377677, supported by cDNA:
gi_20259098 [Arabidopsis thaliana]
gi|18377678|gb|AAL66989.1| putative E2,
ubiquitin-conjugating enzyme [Arabidopsis thaliana]
gi|20259099|gb|AAM14265.1| putative ubiquitin-conjugating
enzyme E2 [Arabidopsis thaliana]
Length = 907
Score = 94.0 bits (232), Expect = 2e-18
Identities = 45/86 (52%), Positives = 59/86 (68%)
Frame = -3
Query: 681 MLYLLRSPPMHFEALVEEHFRQRSKQILLACEAYLEGAPIGYAFDDDGKTEHGNLKGAST 502
M+ +LR PP HFE LV++HF R++ +L AC+AY+EG P+G GN ST
Sbjct: 821 MISMLRKPPKHFEMLVKDHFTHRAQHVLAACKAYMEGVPVG-----SSANLQGNSTTNST 875
Query: 501 GFKIMLAKLLPKLVEAFSDKGIDCHQ 424
GFKIML+KL PKL+EAFS+ G+DC Q
Sbjct: 876 GFKIMLSKLYPKLLEAFSEIGVDCVQ 901
>ref|NP_179284.1| putative ubiquitin-conjugating enzyme; protein id: At2g16920.1
[Arabidopsis thaliana] gi|25411753|pir||H84545 probable
ubiquitin-conjugating enzyme [imported] - Arabidopsis
thaliana gi|3757521|gb|AAC64223.1| putative
ubiquitin-conjugating enzyme [Arabidopsis thaliana]
Length = 1102
Score = 87.0 bits (214), Expect = 2e-16
Identities = 42/93 (45%), Positives = 59/93 (63%), Gaps = 1/93 (1%)
Frame = -3
Query: 681 MLYLLRSPPMHFEALVEEHFRQRSKQILLACEAYLEGAPIGYAFDDDGK-TEHGNLKGAS 505
M+YL+R PP FE L+++HFR+R IL AC+AY++G IG D E + S
Sbjct: 1009 MMYLMRKPPKDFEELIKDHFRKRGYYILKACDAYMKGYLIGSLTKDASVIDERSSANSTS 1068
Query: 504 TGFKIMLAKLLPKLVEAFSDKGIDCHQFAEMQK 406
GFK+MLAK+ PKL A S+ G DC++F +Q+
Sbjct: 1069 VGFKLMLAKIAPKLFSALSEVGADCNEFQHLQQ 1101
>dbj|BAB90019.1| putative ubiquitin-conjugating protein [Oryza sativa (japonica
cultivar-group)]
Length = 493
Score = 73.9 bits (180), Expect = 2e-12
Identities = 37/87 (42%), Positives = 50/87 (56%)
Frame = -3
Query: 681 MLYLLRSPPMHFEALVEEHFRQRSKQILLACEAYLEGAPIGYAFDDDGKTEHGNLKGAST 502
MLY LR+PP HF+ + HF + IL+ C AY++GA +G + GN KG ST
Sbjct: 400 MLYSLRNPPKHFDDFIIGHFHKYGHSILIGCNAYMDGAQVGSIIGGVKAIDKGN-KGCST 458
Query: 501 GFKIMLAKLLPKLVEAFSDKGIDCHQF 421
FK L KL +L+ F G+DCH+F
Sbjct: 459 KFKGSLKKLFEELMMEFIGIGVDCHEF 485
>dbj|BAA96583.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 540
Score = 57.8 bits (138), Expect = 1e-07
Identities = 29/60 (48%), Positives = 37/60 (61%)
Frame = -3
Query: 681 MLYLLRSPPMHFEALVEEHFRQRSKQILLACEAYLEGAPIGYAFDDDGKTEHGNLKGAST 502
MLY LR PP HF LV HFR+R IL AC Y+EG +G ++ + E+G+ GAST
Sbjct: 397 MLYSLRRPPEHFADLVTSHFRERGHTILAACRYYMEGHKVGSVVPEEKEPEYGD-AGAST 455
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 631,100,765
Number of Sequences: 1393205
Number of extensions: 14711646
Number of successful extensions: 39532
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 37739
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39461
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)