Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001339A_C01 KMC001339A_c01
(784 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
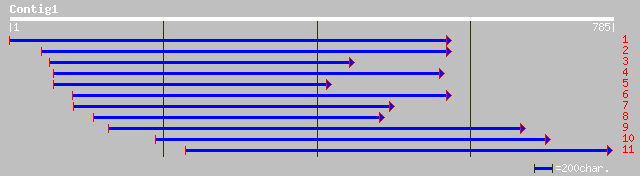
ref|NP_195606.1| kinesin like protein; protein id: At4g38950.1 [... 106 3e-22
gb|AAK91822.1|AF272759_1 kinesin heavy chain [Zea mays] 97 3e-19
gb|AAK91812.1|AF272749_1 kinesin heavy chain [Zea mays] 85 1e-15
dbj|BAB32972.1| putative kinesin [Oryza sativa (japonica cultiva... 77 4e-13
gb|AAF25984.1|AC013354_3 F15H18.12 [Arabidopsis thaliana] 76 6e-13
>ref|NP_195606.1| kinesin like protein; protein id: At4g38950.1 [Arabidopsis thaliana]
gi|7485797|pir||T06055 hypothetical protein F19H22.50 -
Arabidopsis thaliana gi|4539314|emb|CAB38815.1| kinesin
like protein [Arabidopsis thaliana]
gi|7270878|emb|CAB80558.1| kinesin like protein
[Arabidopsis thaliana]
Length = 834
Score = 106 bits (265), Expect = 3e-22
Identities = 54/88 (61%), Positives = 69/88 (78%), Gaps = 3/88 (3%)
Frame = -3
Query: 782 SEEERKRLFKEWDIPLNSKRRRMQLANRLWSNT-DMNHIMQSATVVAKLVRFSEQGKALK 606
S EERKR++ ++ I +NSKRRR+QL N LWSN DM +M+SA VVAKLVRF+EQG+A+K
Sbjct: 746 SGEERKRIYHKFGIAINSKRRRLQLVNELWSNPKDMTQVMESADVVAKLVRFAEQGRAMK 805
Query: 605 EMFGLSFTPQ--LTRRRSYSWKNGRASL 528
EMFGL+FTP LT RRS+SW+ +L
Sbjct: 806 EMFGLTFTPPSFLTTRRSHSWRKSMPAL 833
>gb|AAK91822.1|AF272759_1 kinesin heavy chain [Zea mays]
Length = 897
Score = 96.7 bits (239), Expect = 3e-19
Identities = 46/85 (54%), Positives = 67/85 (78%), Gaps = 2/85 (2%)
Frame = -3
Query: 776 EERKRLFKEWDIPLNSKRRRMQLANRLWSNT-DMNHIMQSATVVAKLVRFSEQGKALKEM 600
+ER+ L+ +W + L+SKRRR+Q+A RLW+ T D+ H+ +SA++VA+L+ E GKAL+EM
Sbjct: 812 QERESLYTKWGVSLSSKRRRLQVARRLWTETRDLEHVRESASLVARLIGLLEPGKALREM 871
Query: 599 FGLSFTPQLTRRRSY-SWKNGRASL 528
FGLSF PQ + RRS+ SW+ GR+SL
Sbjct: 872 FGLSFAPQQSTRRSHSSWRYGRSSL 896
>gb|AAK91812.1|AF272749_1 kinesin heavy chain [Zea mays]
Length = 766
Score = 84.7 bits (208), Expect = 1e-15
Identities = 41/84 (48%), Positives = 59/84 (69%), Gaps = 5/84 (5%)
Frame = -3
Query: 776 EERKRLFKEWDIPLNSKRRRMQLANRLWSNT-DMNHIMQSATVVAKLVRFSEQGKALKEM 600
EER+RL+ +W I L+SK+R++Q+A RLW+ D+ H+ +SA++VAKL+ E G+ L+EM
Sbjct: 678 EERERLYTKWGISLDSKKRKLQVARRLWTEAEDLEHVRESASLVAKLIGLQEPGQVLREM 737
Query: 599 FGLSFTPQL----TRRRSYSWKNG 540
FGLSF PQ RR S W+ G
Sbjct: 738 FGLSFAPQQQPPPRRRSSNGWRYG 761
>dbj|BAB32972.1| putative kinesin [Oryza sativa (japonica cultivar-group)]
Length = 954
Score = 76.6 bits (187), Expect = 4e-13
Identities = 37/74 (50%), Positives = 53/74 (71%), Gaps = 1/74 (1%)
Frame = -3
Query: 782 SEEERKRLFKEWDIPLNSKRRRMQLANRLWSN-TDMNHIMQSATVVAKLVRFSEQGKALK 606
+EEER+RLF +W +PL +K+R++QL NRLW++ D HI +SA +VA+LV F E G K
Sbjct: 861 TEEERERLFIKWQVPLEAKQRKLQLVNRLWTDPNDQAHIDESADIVARLVGFCEGGNISK 920
Query: 605 EMFGLSFTPQLTRR 564
EMF L+F +R+
Sbjct: 921 EMFELNFAVPASRK 934
>gb|AAF25984.1|AC013354_3 F15H18.12 [Arabidopsis thaliana]
Length = 1003
Score = 75.9 bits (185), Expect = 6e-13
Identities = 36/80 (45%), Positives = 52/80 (65%), Gaps = 2/80 (2%)
Frame = -3
Query: 776 EERKRLFKEWDIPLNSKRRRMQLANRLWSNT-DMNHIMQSATVVAKLVRFSEQGKALKEM 600
EER+ L+ +WD+PL K+R++Q N+LW++ D H+ +SA +VAKLV F E G KEM
Sbjct: 917 EEREELYMKWDVPLEGKQRKLQFVNKLWTDPYDSRHVQESAEIVAKLVGFCESGNISKEM 976
Query: 599 FGLSF-TPQLTRRRSYSWKN 543
F L+F P R+ + W N
Sbjct: 977 FELNFAVPSDKRQWNIGWDN 996
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 662,650,775
Number of Sequences: 1393205
Number of extensions: 14227494
Number of successful extensions: 40370
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 38874
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40340
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38935490438
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)