Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001338A_C01 KMC001338A_c01
(825 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
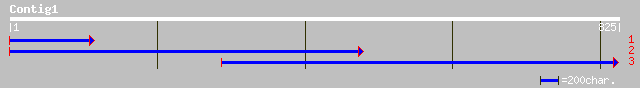
dbj|BAC41798.1| unknown protein [Arabidopsis thaliana] gi|290290... 41 0.019
ref|NP_182303.1| hypothetical protein; protein id: At2g47820.1 [... 41 0.019
dbj|BAC55669.1| OJ1092_A07.23 [Oryza sativa (japonica cultivar-g... 36 0.61
ref|NP_700656.1| hypothetical protein [Plasmodium falciparum 3D7... 32 8.7
>dbj|BAC41798.1| unknown protein [Arabidopsis thaliana] gi|29029062|gb|AAO64910.1|
At2g47820 [Arabidopsis thaliana]
Length = 805
Score = 41.2 bits (95), Expect = 0.019
Identities = 19/40 (47%), Positives = 24/40 (59%)
Frame = -2
Query: 824 RNRPPTTKVLEAFAFGYLDQKEKRKRKDIQDRSVSRPSRR 705
R RP TTK LEAFAFGYL K ++ + R+ S R+
Sbjct: 728 RTRPLTTKALEAFAFGYLGNSNKERKASEESRTKSNKKRK 767
>ref|NP_182303.1| hypothetical protein; protein id: At2g47820.1 [Arabidopsis
thaliana] gi|25370841|pir||H84919 hypothetical protein
At2g47820 [imported] - Arabidopsis thaliana
gi|3738294|gb|AAC63636.1| hypothetical protein
[Arabidopsis thaliana]
Length = 797
Score = 41.2 bits (95), Expect = 0.019
Identities = 19/40 (47%), Positives = 24/40 (59%)
Frame = -2
Query: 824 RNRPPTTKVLEAFAFGYLDQKEKRKRKDIQDRSVSRPSRR 705
R RP TTK LEAFAFGYL K ++ + R+ S R+
Sbjct: 720 RTRPLTTKALEAFAFGYLGNSNKERKASEESRTKSNKKRK 759
>dbj|BAC55669.1| OJ1092_A07.23 [Oryza sativa (japonica cultivar-group)]
Length = 1031
Score = 36.2 bits (82), Expect = 0.61
Identities = 20/44 (45%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Frame = -2
Query: 824 RNRPPTTKVLEAFAFGYLDQKEKRKRKDIQDRS-VSRPSRRQCR 696
R+RPPTT+ LEA A G++ K+K + S SRP RR R
Sbjct: 873 RSRPPTTRALEALACGFIGTKQKGAEGNFPSSSRSSRPVRRPRR 916
>ref|NP_700656.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23495048|gb|AAN35380.1|AE014832_2 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 716
Score = 32.3 bits (72), Expect = 8.7
Identities = 17/59 (28%), Positives = 28/59 (46%)
Frame = -2
Query: 764 KEKRKRKDIQDRSVSRPSRRQCRKEVVGSSSDCAGFEKEETRNVCNGHGNGSTGSHSDV 588
++K+K+KD ++ S C K + KE +N CN + N +T HS+V
Sbjct: 303 QKKKKKKDYDKHNIYNESSNVCDKYI-----------KEVLQNNCNSYNNNNTYDHSNV 350
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 677,870,647
Number of Sequences: 1393205
Number of extensions: 14228863
Number of successful extensions: 31841
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 30566
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31784
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42857050626
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)