Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001325A_C01 KMC001325A_c01
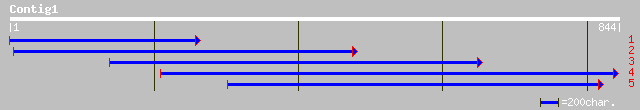
(772 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||C84781 hypothetical protein At2g36480 [imported] - Arabidop... 64 3e-09
ref|NP_565848.1| hypothetical protein; protein id: At2g36480.1 [... 64 3e-09
ref|NP_680598.1| similar to S-locus protein 4-like; protein id: ... 55 1e-06
gb|AAD03447.1| contains similarity to human PCF11p homolog (GB:A... 55 1e-06
gb|AAL57625.1| At1g66500/F28G11_6 [Arabidopsis thaliana] 45 0.002
>pir||C84781 hypothetical protein At2g36480 [imported] - Arabidopsis thaliana
Length = 984
Score = 63.5 bits (153), Expect = 3e-09
Identities = 27/64 (42%), Positives = 39/64 (60%)
Frame = -2
Query: 771 CLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLSENP 592
C C+LCGE+FE+++ QE +WMFKGA Y+ N N S GPI+H CL+ +
Sbjct: 904 CACILCGEVFEDYFSQEMAQWMFKGASYLTNPPAN--------SEASGPIVHTGCLTTSS 955
Query: 591 VSSV 580
+ S+
Sbjct: 956 LQSL 959
>ref|NP_565848.1| hypothetical protein; protein id: At2g36480.1 [Arabidopsis
thaliana] gi|20197927|gb|AAD24632.2| hypothetical
protein [Arabidopsis thaliana]
Length = 828
Score = 63.5 bits (153), Expect = 3e-09
Identities = 27/64 (42%), Positives = 39/64 (60%)
Frame = -2
Query: 771 CLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLSENP 592
C C+LCGE+FE+++ QE +WMFKGA Y+ N N S GPI+H CL+ +
Sbjct: 761 CACILCGEVFEDYFSQEMAQWMFKGASYLTNPPAN--------SEASGPIVHTGCLTTSS 812
Query: 591 VSSV 580
+ S+
Sbjct: 813 LQSL 816
>ref|NP_680598.1| similar to S-locus protein 4-like; protein id: At4g04885.1
[Arabidopsis thaliana]
Length = 831
Score = 54.7 bits (130), Expect = 1e-06
Identities = 29/59 (49%), Positives = 35/59 (59%), Gaps = 2/59 (3%)
Frame = -2
Query: 765 CVLCGELFEEFYCQENGEWMFKGAVYV--PNSDINDDMGIRDASTGGGPIIHARCLSEN 595
C LCGE FE+FY E EWM+KGAVY+ P D D S GPI+HA+C E+
Sbjct: 762 CALCGEPFEDFYSDETEEWMYKGAVYMNAPEESTTD----MDKSQ-LGPIVHAKCRPES 815
>gb|AAD03447.1| contains similarity to human PCF11p homolog (GB:AF046935)
[Arabidopsis thaliana]
Length = 827
Score = 54.7 bits (130), Expect = 1e-06
Identities = 29/59 (49%), Positives = 35/59 (59%), Gaps = 2/59 (3%)
Frame = -2
Query: 765 CVLCGELFEEFYCQENGEWMFKGAVYV--PNSDINDDMGIRDASTGGGPIIHARCLSEN 595
C LCGE FE+FY E EWM+KGAVY+ P D D S GPI+HA+C E+
Sbjct: 762 CALCGEPFEDFYSDETEEWMYKGAVYMNAPEESTTD----MDKSQ-LGPIVHAKCRPES 815
>gb|AAL57625.1| At1g66500/F28G11_6 [Arabidopsis thaliana]
Length = 416
Score = 44.7 bits (104), Expect = 0.002
Identities = 21/56 (37%), Positives = 29/56 (51%)
Frame = -2
Query: 765 CVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLSE 598
C LC E FEEF+ E+ +WM+K AVY+ T G I+H +C+ E
Sbjct: 349 CALCVEPFEEFFSHEDDDWMYKDAVYL---------------TKNGRIVHVKCMPE 389
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 639,826,150
Number of Sequences: 1393205
Number of extensions: 13698783
Number of successful extensions: 52437
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 43230
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 50732
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37815044670
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)