Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001314A_C01 KMC001314A_c01
(791 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
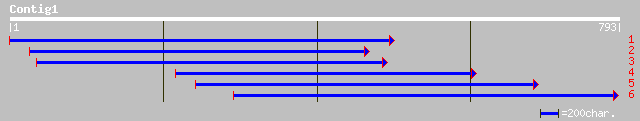
ref|NP_171966.1| hypothetical protein; protein id: At1g04730.1 [... 67 2e-10
pir||B86180 protein T1G11.3 [imported] - Arabidopsis thaliana gi... 36 0.73
gb|AAO50743.1| similar to hypothetical protein; protein id: At1g... 36 0.73
ref|NP_010792.1| Hypothetical ORF; Ydr504cp [Saccharomyces cerev... 34 2.1
sp|P45481|CBP_MOUSE CREB-binding protein gi|481698|pir||S39161 C... 33 6.2
>ref|NP_171966.1| hypothetical protein; protein id: At1g04730.1 [Arabidopsis thaliana]
Length = 954
Score = 67.4 bits (163), Expect = 2e-10
Identities = 38/81 (46%), Positives = 47/81 (57%)
Frame = -1
Query: 782 PNGSANATDXAKLLKMGKLKKPSSSSSFFDRFKKLNVKGLPNDRSQQKEATLEKHRCPLL 603
P+ S +K K ++ +FFDRF+K K + Q AT ++ PLL
Sbjct: 874 PSVSVGTATTSKPNSSDVKKASRNALNFFDRFRKSR-KDYEDPEDVQNRATAKRDSRPLL 932
Query: 602 FKFNEGFTNAVKRPVRMREFL 540
FKFNEGFTNAVKRPVRMREFL
Sbjct: 933 FKFNEGFTNAVKRPVRMREFL 953
>pir||B86180 protein T1G11.3 [imported] - Arabidopsis thaliana
gi|2494129|gb|AAB80638.1| T1G11.3 [Arabidopsis thaliana]
Length = 443
Score = 35.8 bits (81), Expect = 0.73
Identities = 22/65 (33%), Positives = 31/65 (46%)
Frame = -1
Query: 782 PNGSANATDXAKLLKMGKLKKPSSSSSFFDRFKKLNVKGLPNDRSQQKEATLEKHRCPLL 603
P+ S +K K ++ +FFDRF+K K + Q AT ++ PLL
Sbjct: 365 PSVSVGTATTSKPNSSDVKKASRNALNFFDRFRKSR-KDYEDPEDVQNRATAKRDSRPLL 423
Query: 602 FKFNE 588
FKFNE
Sbjct: 424 FKFNE 428
>gb|AAO50743.1| similar to hypothetical protein; protein id: At1g04730.1 [Arabidopsis
thaliana] [Dictyostelium discoideum]
Length = 937
Score = 35.8 bits (81), Expect = 0.73
Identities = 21/64 (32%), Positives = 37/64 (57%)
Frame = -1
Query: 731 KLKKPSSSSSFFDRFKKLNVKGLPNDRSQQKEATLEKHRCPLLFKFNEGFTNAVKRPVRM 552
+ KKP + D F ++ ++ P + + EAT + + ++F EGFTNAVK+ V +
Sbjct: 878 EFKKPLVPTVLKDFFGRV-IQTSPETKKKTMEATAGPN---IKYRFQEGFTNAVKKTVYV 933
Query: 551 REFL 540
++FL
Sbjct: 934 KDFL 937
>ref|NP_010792.1| Hypothetical ORF; Ydr504cp [Saccharomyces cerevisiae]
gi|2131535|pir||S69562 hypothetical protein YDR504c -
yeast (Saccharomyces cerevisiae)
gi|927774|gb|AAB64946.1| Ydr504cp; CAI: 0.21
[Saccharomyces cerevisiae]
Length = 127
Score = 34.3 bits (77), Expect = 2.1
Identities = 22/52 (42%), Positives = 27/52 (51%), Gaps = 6/52 (11%)
Frame = -2
Query: 607 YFLNLMRVLLMLSKGLFEC--VNFFPNYAFNK*CSLFTYLP----WYYRFII 470
YF+N+ + L L +F V FF F + CSLFTY P WYY II
Sbjct: 22 YFVNIFSLFLFLFVFVFVFIFVYFFYVILFYRFCSLFTYFPANSIWYYLSII 73
>sp|P45481|CBP_MOUSE CREB-binding protein gi|481698|pir||S39161 CREB-binding protein -
mouse gi|435855|gb|AAB28651.1| CREB-binding protein; CBP
[Mus sp.]
Length = 2441
Score = 32.7 bits (73), Expect = 6.2
Identities = 21/59 (35%), Positives = 31/59 (51%)
Frame = -1
Query: 791 NSNPNGSANATDXAKLLKMGKLKKPSSSSSFFDRFKKLNVKGLPNDRSQQKEATLEKHR 615
+ P GS + AK K K SS S ++ KK ++ + ND SQ+ AT+EKH+
Sbjct: 1573 SETPEGSQGDSKNAKKKNNKKTNKNKSSISRANK-KKPSMPNVSNDLSQKLYATMEKHK 1630
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 597,814,769
Number of Sequences: 1393205
Number of extensions: 11940866
Number of successful extensions: 23433
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 22917
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23426
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 39775824764
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)