Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001299A_C01 KMC001299A_c01
(554 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
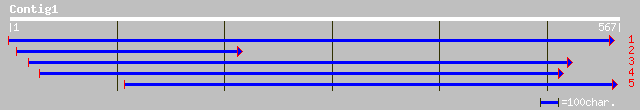
ref|NP_199933.1| putative protein; protein id: At5g51200.1 [Arab... 115 4e-25
gb|AAF81925.1|AF107287_1 elongation factor 2 [Candida glabrata] 33 3.0
ref|NP_190628.1| hypothetical protein; protein id: At3g50590.1 [... 33 3.0
ref|NP_199827.2| FRO1 and FRO2-like protein; protein id: At5g501... 32 4.0
dbj|BAB09387.1| FRO1 and FRO2-like protein [Arabidopsis thaliana] 32 4.0
>ref|NP_199933.1| putative protein; protein id: At5g51200.1 [Arabidopsis thaliana]
gi|8843857|dbj|BAA97383.1| contains similarity to unknown
protein~dbj|BAA13214.1~gene_id:MWD22.15 [Arabidopsis
thaliana]
Length = 1837
Score = 115 bits (288), Expect = 4e-25
Identities = 67/148 (45%), Positives = 90/148 (60%), Gaps = 28/148 (18%)
Frame = -2
Query: 553 LSSADNIPKRRYLAMVEMCRVVASRDRLIILLLPLLEHVLNIILIHLQNSSIALDPAMST 374
++ +DNI KRRY+AMVEMC++V +RD+LI LLL L EHVLNIILIHLQ+ S++ S
Sbjct: 1695 VTPSDNIHKRRYIAMVEMCQIVGNRDQLITLLLQLAEHVLNIILIHLQDRSVS-----SN 1749
Query: 373 KTITYSSKCDPQQDMALFCGKLVPTLERLEL----------------------------L 278
+ +Y SK QQ++ CGKL PT++RL L L
Sbjct: 1750 ERGSYGSKSHIQQEVTDLCGKLSPTIDRLALLNEVSLIDFREFSIIYVLKVKGGGDPICL 1809
Query: 277 SEEKIGHNLKVFPRLTTSAQEIAIPKLI 194
S+ K+GHNLKVF RL T+ +E+AI K +
Sbjct: 1810 SQGKVGHNLKVFQRLATTVKEMAIQKCV 1837
>gb|AAF81925.1|AF107287_1 elongation factor 2 [Candida glabrata]
Length = 814
Score = 32.7 bits (73), Expect = 3.0
Identities = 20/65 (30%), Positives = 32/65 (48%), Gaps = 9/65 (13%)
Frame = -2
Query: 457 LPLLEHVLNIILIHLQNSSIALD---------PAMSTKTITYSSKCDPQQDMALFCGKLV 305
LP + +L +I++HL + A + PA I KCDP D+ L+ K+V
Sbjct: 314 LPAADALLEMIVMHLPSPVTAQNYRAEQLYEGPADDANCIAIK-KCDPTADLMLYVSKMV 372
Query: 304 PTLER 290
PT ++
Sbjct: 373 PTSDK 377
>ref|NP_190628.1| hypothetical protein; protein id: At3g50590.1 [Arabidopsis
thaliana] gi|11280713|pir||T46090 hypothetical protein
T20E23.190 - Arabidopsis thaliana
gi|6561999|emb|CAB62488.1| hypothetical protein
[Arabidopsis thaliana]
Length = 690
Score = 32.7 bits (73), Expect = 3.0
Identities = 28/113 (24%), Positives = 52/113 (45%), Gaps = 5/113 (4%)
Frame = -2
Query: 550 SSADNIPKRRYLAM-----VEMCRVVASRDRLIILLLPLLEHVLNIILIHLQNSSIALDP 386
S DNI K+ Y ++ +V SR R + L++ N ++++ +N+ L
Sbjct: 517 SGTDNIYKKLYTSIPGNVEYHSKHIVYSRKRHLFLVVFEFSGATNEVVLYWENTGSQLPN 576
Query: 385 AMSTKTITYSSKCDPQQDMALFCGKLVPTLERLELLSEEKIGHNLKVFPRLTT 227
+ + + CD A F G P ++ +L E+K G ++ + P+LTT
Sbjct: 577 SKGST----AKGCD-----AAFIG---PNDDQFAILDEDKTGLSMYILPKLTT 617
>ref|NP_199827.2| FRO1 and FRO2-like protein; protein id: At5g50160.1, supported by
cDNA: gi_18377667 [Arabidopsis thaliana]
gi|18377668|gb|AAL66984.1| putative FRO1 and FRO2
protein [Arabidopsis thaliana]
gi|27754744|gb|AAO22815.1| putative FRO1 and FRO2
protein [Arabidopsis thaliana]
Length = 728
Score = 32.3 bits (72), Expect = 4.0
Identities = 22/77 (28%), Positives = 39/77 (50%)
Frame = +3
Query: 21 KVHNLNKFSPLHYKISLHICKFKLIQKEHLHFIQFTVYRQLIQSRQLNLYWIL*YIYKIN 200
K NLN + +Y+++ +F L+ + L + F V R L R LN+ + Y +
Sbjct: 141 KTMNLNLWQLKYYRVAT---RFGLLAEACLSLLLFPVLRGLSMFRLLNIEFAASVKYHVW 197
Query: 201 FGIAISWAEVVNLGNTL 251
FG + + +V+ G+TL
Sbjct: 198 FGTGLIFFSLVHGGSTL 214
>dbj|BAB09387.1| FRO1 and FRO2-like protein [Arabidopsis thaliana]
Length = 713
Score = 32.3 bits (72), Expect = 4.0
Identities = 22/77 (28%), Positives = 39/77 (50%)
Frame = +3
Query: 21 KVHNLNKFSPLHYKISLHICKFKLIQKEHLHFIQFTVYRQLIQSRQLNLYWIL*YIYKIN 200
K NLN + +Y+++ +F L+ + L + F V R L R LN+ + Y +
Sbjct: 141 KTMNLNLWQLKYYRVAT---RFGLLAEACLSLLLFPVLRGLSMFRLLNIEFAASVKYHVW 197
Query: 201 FGIAISWAEVVNLGNTL 251
FG + + +V+ G+TL
Sbjct: 198 FGTGLIFFSLVHGGSTL 214
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 455,260,840
Number of Sequences: 1393205
Number of extensions: 9226136
Number of successful extensions: 33340
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 32744
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33337
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)