Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001293A_C01 KMC001293A_c01
(954 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
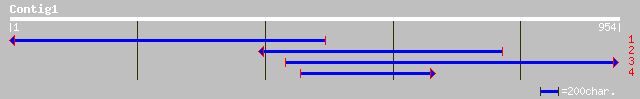
ref|NP_565266.1| expressed protein; protein id: At2g01590.1, sup... 107 2e-22
gb|AAM18505.1|AF494415_1 N-methyltransferase [Coffea canephora] 78 2e-13
gb|AAF98284.1|AF198492_1 SAM:benzoic acid carboxyl methyltransfe... 78 2e-13
gb|AAM18506.1|AF494416_1 N-methyltransferase [Coffea canephora] 78 2e-13
dbj|BAB39215.1| theobromine synthase [Coffea arabica] gi|2027102... 78 2e-13
>ref|NP_565266.1| expressed protein; protein id: At2g01590.1, supported by cDNA:
gi_14334561, supported by cDNA: gi_21281045 [Arabidopsis
thaliana] gi|25371191|pir||F84426 hypothetical protein
At2g01590 [imported] - Arabidopsis thaliana
gi|3785988|gb|AAC67335.1| expressed protein [Arabidopsis
thaliana] gi|14334562|gb|AAK59689.1| unknown protein
[Arabidopsis thaliana] gi|21281046|gb|AAM44943.1|
unknown protein [Arabidopsis thaliana]
Length = 174
Score = 107 bits (268), Expect = 2e-22
Identities = 56/115 (48%), Positives = 77/115 (66%), Gaps = 2/115 (1%)
Frame = +2
Query: 38 RRRLPQPSVIRMERIVGTGSFRDGEPEPRDSDVRKTIFDLFLGQA--FEGPVEKKLRETG 211
+RR +PS+ ++ER G+GS+RD E E D+ +L LG A FE +EKKLRE G
Sbjct: 63 QRRGNKPSIAQIERAFGSGSYRDSEGE---MDMNTVFDELLLGHANKFESKIEKKLREIG 119
Query: 212 EWVAANSEARLRSAGKGTLMFMFVWMLPIWAILLILATGTIKLPFSSPFLDEILL 376
E A +E +LRS+GK LMF W+LPIW + L++A G IKLPFS PFLD++++
Sbjct: 120 EIFVARTEPKLRSSGKPVLMFTIQWILPIWIMSLLVACGVIKLPFSIPFLDDLIM 174
>gb|AAM18505.1|AF494415_1 N-methyltransferase [Coffea canephora]
Length = 371
Score = 78.2 bits (191), Expect = 2e-13
Identities = 41/112 (36%), Positives = 67/112 (59%)
Frame = -1
Query: 954 LDYFNMPLYCPTMEEVKXIIEGEGSFTLQTLKTIQISWNSHLPDDIDVSVLDNXMRGYLI 775
LD FN+P+Y P+ EEVK I+E EGSF + L+T ++ + D S+ D ++ +
Sbjct: 259 LDSFNLPVYIPSAEEVKCIVEEEGSFEILYLETFKVLY------DAGFSIDDEHIKAEYV 312
Query: 774 AKSLRAVMEPLLSFAFGNNIMDELFSRFGKKISQIMEMETLQYTTLIMYMTK 619
A S+RAV EP+L+ FG I+ ++F RF K ++++ + Y LI+ + K
Sbjct: 313 ASSVRAVYEPILASHFGEAIIPDIFHRFAKHAAKVLPLGKGFYNNLIISLAK 364
>gb|AAF98284.1|AF198492_1 SAM:benzoic acid carboxyl methyltransferase [Antirrhinum majus]
Length = 364
Score = 78.2 bits (191), Expect = 2e-13
Identities = 39/113 (34%), Positives = 67/113 (58%), Gaps = 4/113 (3%)
Frame = -1
Query: 945 FNMPLYCPTMEEVKXIIEGEGSFTLQTLKTIQISWNS--HLPDDI--DVSVLDNXMRGYL 778
FN+P+Y P EV+ I EGSFTL L+ ++ W++ + DD D S+ G
Sbjct: 251 FNIPIYSPCTREVEAAILSEGSFTLDRLEVFRVCWDASDYTDDDDQQDPSIFGKQRSGKF 310
Query: 777 IAKSLRAVMEPLLSFAFGNNIMDELFSRFGKKISQIMEMETLQYTTLIMYMTK 619
+A +RA+ EP+L+ FG+ IMD LF ++ KKI + + +E Y ++++ +++
Sbjct: 311 VADCVRAITEPMLASHFGSTIMDLLFGKYAKKIVEHLSVENSSYFSIVVSLSR 363
>gb|AAM18506.1|AF494416_1 N-methyltransferase [Coffea canephora]
Length = 372
Score = 78.2 bits (191), Expect = 2e-13
Identities = 41/112 (36%), Positives = 67/112 (59%)
Frame = -1
Query: 954 LDYFNMPLYCPTMEEVKXIIEGEGSFTLQTLKTIQISWNSHLPDDIDVSVLDNXMRGYLI 775
LD FN+P+Y P+ EEVK I+E EGSF + L+T ++ + D S+ D ++ +
Sbjct: 260 LDSFNLPVYIPSAEEVKCIVEEEGSFEILYLETFKVLY------DAGFSIDDEHIKAEYV 313
Query: 774 AKSLRAVMEPLLSFAFGNNIMDELFSRFGKKISQIMEMETLQYTTLIMYMTK 619
A S+RAV EP+L+ FG I+ ++F RF K ++++ + Y LI+ + K
Sbjct: 314 ASSVRAVYEPILASHFGEAIIPDIFHRFAKHAAKVLPLGKGFYNNLIISLAK 365
>dbj|BAB39215.1| theobromine synthase [Coffea arabica]
gi|20271020|gb|AAM18502.1|AF494412_1 N-methyltransferase
[Coffea arabica] gi|26453373|dbj|BAC43755.1| tentative
caffeine synthase 1 [Coffea arabica]
Length = 372
Score = 78.2 bits (191), Expect = 2e-13
Identities = 41/112 (36%), Positives = 67/112 (59%)
Frame = -1
Query: 954 LDYFNMPLYCPTMEEVKXIIEGEGSFTLQTLKTIQISWNSHLPDDIDVSVLDNXMRGYLI 775
LD FN+P+Y P+ EEVK I+E EGSF + L+T ++ + D S+ D ++ +
Sbjct: 260 LDSFNLPVYIPSAEEVKCIVEEEGSFEILYLETFKVLY------DAGFSIDDEHIKAEYV 313
Query: 774 AKSLRAVMEPLLSFAFGNNIMDELFSRFGKKISQIMEMETLQYTTLIMYMTK 619
A S+RAV EP+L+ FG I+ ++F RF K ++++ + Y LI+ + K
Sbjct: 314 ASSVRAVYEPILASHFGEAIIPDIFHRFAKHAAKVLPLGKGFYNNLIISLAK 365
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 829,782,954
Number of Sequences: 1393205
Number of extensions: 18453040
Number of successful extensions: 53469
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 50723
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 53337
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 53801056208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)