Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001283A_C01 KMC001283A_c01
(818 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
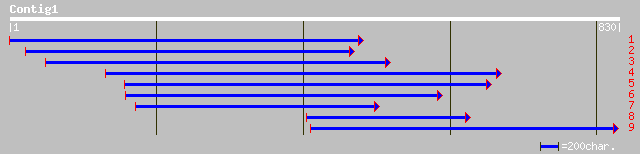
sp|P53537|PHSH_VICFA ALPHA-GLUCAN PHOSPHORYLASE, H ISOZYME (STAR... 47 4e-04
ref|NP_190281.1| starch phosphorylase, putative; protein id: At3... 44 0.003
gb|AAL10498.1| AT3g46970/F13I12_20 [Arabidopsis thaliana] 44 0.003
sp|P32811|PHSH_SOLTU ALPHA-GLUCAN PHOSPHORYLASE, H ISOZYME (STAR... 44 0.003
gb|AAM29154.1| starch phosphorylase type H [Citrus hybrid cultivar] 43 0.005
>sp|P53537|PHSH_VICFA ALPHA-GLUCAN PHOSPHORYLASE, H ISOZYME (STARCH PHOSPHORYLASE H)
gi|7433824|pir||T12091 starch phosphorylase (EC 2.4.1.1)
H, cytosolic isoform - fava bean
gi|510932|emb|CAA84494.1| alpha 1,4-glucan phosphorylase
type H [Vicia faba]
Length = 842
Score = 46.6 bits (109), Expect = 4e-04
Identities = 19/35 (54%), Positives = 26/35 (74%)
Frame = -2
Query: 817 QMSILSVAGSGRFTRDRHLPEYAERTWKIDPCRCP 713
+MSILS AGSG+F+ DR + +YA+ W I+ CR P
Sbjct: 808 KMSILSTAGSGKFSSDRTIAQYAKEIWNIEECRVP 842
>ref|NP_190281.1| starch phosphorylase, putative; protein id: At3g46970.1, supported
by cDNA: gi_15983802, supported by cDNA: gi_19699064
[Arabidopsis thaliana] gi|14916634|sp|Q9SD76|PHSH_ARATH
Alpha-glucan phosphorylase, H isozyme (Starch
phosphorylase H) gi|11257928|pir||T45633 starch
phosphorylase (EC 2.4.1.1) H, cytosolic F13I12.20
[similarity] - Arabidopsis thaliana
gi|6522578|emb|CAB61943.1| starch phosphorylase H
(cytosolic form)-like protein [Arabidopsis thaliana]
gi|19699065|gb|AAL90900.1| AT3g46970/F13I12_20
[Arabidopsis thaliana] gi|27764912|gb|AAO23577.1|
At3g46970/F13I12_20 [Arabidopsis thaliana]
Length = 841
Score = 43.9 bits (102), Expect = 0.003
Identities = 18/35 (51%), Positives = 25/35 (71%)
Frame = -2
Query: 817 QMSILSVAGSGRFTRDRHLPEYAERTWKIDPCRCP 713
+MSILS AGSG+F+ DR + +YA+ W I+ C P
Sbjct: 807 KMSILSTAGSGKFSSDRTIAQYAKEIWNIEACPVP 841
>gb|AAL10498.1| AT3g46970/F13I12_20 [Arabidopsis thaliana]
Length = 841
Score = 43.9 bits (102), Expect = 0.003
Identities = 18/35 (51%), Positives = 25/35 (71%)
Frame = -2
Query: 817 QMSILSVAGSGRFTRDRHLPEYAERTWKIDPCRCP 713
+MSILS AGSG+F+ DR + +YA+ W I+ C P
Sbjct: 807 KMSILSTAGSGKFSSDRTIAQYAKEIWNIEACPVP 841
>sp|P32811|PHSH_SOLTU ALPHA-GLUCAN PHOSPHORYLASE, H ISOZYME (STARCH PHOSPHORYLASE H)
gi|100452|pir||A40995 starch phosphorylase (EC 2.4.1.1)
H [similarity] - potato gi|169473|gb|AAA33809.1|
alpha-glucan phosphorylase type H isozyme
Length = 838
Score = 43.9 bits (102), Expect = 0.003
Identities = 18/35 (51%), Positives = 25/35 (71%)
Frame = -2
Query: 817 QMSILSVAGSGRFTRDRHLPEYAERTWKIDPCRCP 713
+MSILS +GSG+F+ DR + +YA+ W I CR P
Sbjct: 804 KMSILSTSGSGKFSSDRTISQYAKEIWNIAECRVP 838
>gb|AAM29154.1| starch phosphorylase type H [Citrus hybrid cultivar]
Length = 840
Score = 43.1 bits (100), Expect = 0.005
Identities = 18/33 (54%), Positives = 24/33 (72%)
Frame = -2
Query: 817 QMSILSVAGSGRFTRDRHLPEYAERTWKIDPCR 719
+MSILS AGSG+F+ DR + +YA+ W I CR
Sbjct: 806 KMSILSTAGSGKFSSDRTIAQYAKEIWNITECR 838
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 690,509,779
Number of Sequences: 1393205
Number of extensions: 15109697
Number of successful extensions: 34275
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 33003
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34247
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42296827742
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)