Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001278A_C01 KMC001278A_c01
(562 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
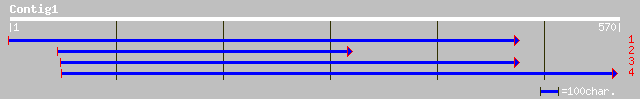
gb|AAO22731.1| unknown protein [Arabidopsis thaliana] 59 4e-08
ref|NP_192387.1| hypothetical protein; protein id: At4g04780.1 [... 59 4e-08
gb|AAD03443.1| contains similarity to human RNA polymerase II co... 50 1e-05
gb|AAL73528.1|AF466200_7 putative RNA polymerase II complex comp... 42 0.007
dbj|BAC29578.1| unnamed protein product [Mus musculus] 33 1.8
>gb|AAO22731.1| unknown protein [Arabidopsis thaliana]
Length = 162
Score = 58.9 bits (141), Expect = 4e-08
Identities = 45/119 (37%), Positives = 54/119 (44%), Gaps = 40/119 (33%)
Frame = -2
Query: 561 MDLISQLKEQVNLTSTDLLKAFKKL------------------------------EQPHQ 472
MD+ISQL+EQVN + AF L EQP Q
Sbjct: 24 MDIISQLQEQVNTIAAITFNAFGTLQRDAPPVQLSPNYPEPPATTTVTDDATPFPEQPKQ 83
Query: 471 ----------QLIALVAALLISEGDEEAQLKRIAELQAENDAIGQEPAEAIGICRQRIK 325
Q ALVAAL +SEG E AQLKRIAELQ END +GQE + + + +K
Sbjct: 84 LSAGLVKAAKQFDALVAALPLSEGGEGAQLKRIAELQVENDLVGQELQKQLEAAEKELK 142
Score = 45.4 bits (106), Expect = 5e-04
Identities = 22/35 (62%), Positives = 28/35 (79%)
Frame = -3
Query: 368 KNLQKLLEFADKELNQIQELFIQASDIYLNLKKPD 264
+ LQK LE A+KEL Q+QELF QA+D LN+KKP+
Sbjct: 128 QELQKQLEAAEKELKQVQELFGQAADNCLNMKKPE 162
>ref|NP_192387.1| hypothetical protein; protein id: At4g04780.1 [Arabidopsis
thaliana] gi|25407290|pir||B85060 hypothetical protein
AT4g04780 [imported] - Arabidopsis thaliana
gi|7267236|emb|CAB80843.1| hypothetical protein
[Arabidopsis thaliana]
Length = 381
Score = 58.9 bits (141), Expect = 4e-08
Identities = 45/119 (37%), Positives = 54/119 (44%), Gaps = 40/119 (33%)
Frame = -2
Query: 561 MDLISQLKEQVNLTSTDLLKAFKKL------------------------------EQPHQ 472
MD+ISQL+EQVN + AF L EQP Q
Sbjct: 243 MDIISQLQEQVNTIAAITFNAFGTLQRDAPPVQLSPNYPEPPATTTVTDDATPFPEQPKQ 302
Query: 471 ----------QLIALVAALLISEGDEEAQLKRIAELQAENDAIGQEPAEAIGICRQRIK 325
Q ALVAAL +SEG E AQLKRIAELQ END +GQE + + + +K
Sbjct: 303 LSAGLVKAAKQFDALVAALPLSEGGEGAQLKRIAELQVENDLVGQELQKQLEAAEKELK 361
Score = 45.4 bits (106), Expect = 5e-04
Identities = 22/35 (62%), Positives = 28/35 (79%)
Frame = -3
Query: 368 KNLQKLLEFADKELNQIQELFIQASDIYLNLKKPD 264
+ LQK LE A+KEL Q+QELF QA+D LN+KKP+
Sbjct: 347 QELQKQLEAAEKELKQVQELFGQAADNCLNMKKPE 381
>gb|AAD03443.1| contains similarity to human RNA polymerase II complex component
SRB7 (GB:U52960) [Arabidopsis thaliana]
Length = 168
Score = 50.4 bits (119), Expect = 1e-05
Identities = 46/138 (33%), Positives = 57/138 (40%), Gaps = 49/138 (35%)
Frame = -2
Query: 561 MDLISQLKEQVNLTSTDLLKAFKKL------------------------------EQPHQ 472
MD+ISQL+EQVN + AF L EQP Q
Sbjct: 1 MDIISQLQEQVNTIAAITFNAFGTLQRDAPPVQLSPNYPEPPATTTVTDDATPFPEQPKQ 60
Query: 471 ----------QLIALVAALLISEGDEEAQLKRIAELQ---------AENDAIGQEPAEAI 349
Q ALVAAL +SEG E AQLKRIAELQ END +GQE + +
Sbjct: 61 LSAGLVKAAKQFDALVAALPLSEGGEGAQLKRIAELQVKQVTPICRVENDLVGQELQKQL 120
Query: 348 GICRQRIKSNSGVVYSSI 295
+ + VV + +
Sbjct: 121 EAAEGAVAKVAEVVVAEV 138
>gb|AAL73528.1|AF466200_7 putative RNA polymerase II complex component SRB7 protein [Sorghum
bicolor]
Length = 159
Score = 41.6 bits (96), Expect = 0.007
Identities = 30/73 (41%), Positives = 40/73 (54%)
Frame = -2
Query: 543 LKEQVNLTSTDLLKAFKKLEQPHQQLIALVAALLISEGDEEAQLKRIAELQAENDAIGQE 364
L EQ S L+ A KK + ALVAAL +S EE Q+KRI ELQAEN+ +G E
Sbjct: 76 LSEQPKAMSHALVLAAKKFD-------ALVAALPLSS--EEDQVKRIQELQAENEVVGLE 126
Query: 363 PAEAIGICRQRIK 325
+ + + +K
Sbjct: 127 LQKQLEAAERELK 139
>dbj|BAC29578.1| unnamed protein product [Mus musculus]
Length = 526
Score = 33.5 bits (75), Expect = 1.8
Identities = 34/125 (27%), Positives = 59/125 (47%), Gaps = 15/125 (12%)
Frame = -2
Query: 546 QLKEQVNLTSTDLLKAFKKLEQPHQQLIALVAA--------LLISEGDEEAQLKRIAELQ 391
+L+ Q+ +T L+ ++L+Q + +L +AA + EGD EA ++ E +
Sbjct: 96 ELRRQLETLNTQHLEHEERLQQENHELRRGLAARGAEWEARAVELEGDVEALRAQLGEQR 155
Query: 390 AENDAIGQEPAEAIG-ICRQRIKSNSGVVYSS------IRYLFELEETRCQLLAGVAVEA 232
+E G+E A A+G + Q ++ + + +S R L L E RCQ A E
Sbjct: 156 SERQDSGRERARALGELSEQNLRLSQQLAQASRTEQELQRELDTLRE-RCQTQALAGAEL 214
Query: 231 GDKSE 217
G + E
Sbjct: 215 GARLE 219
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 417,269,006
Number of Sequences: 1393205
Number of extensions: 8210890
Number of successful extensions: 25774
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 24734
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25731
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)