Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001269A_C01 KMC001269A_c01
(564 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
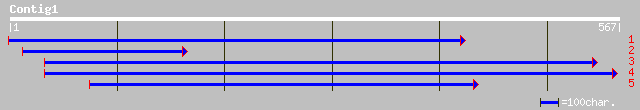
emb|CAB90633.1| protein phpsphatase 2C (PP2C) [Fagus sylvatica] 64 1e-09
emb|CAC09575.1| protein phosphatase 2C (PP2C) [Fagus sylvatica] 62 4e-09
gb|AAM61361.1| protein phosphatase 2C, putative [Arabidopsis tha... 55 5e-07
ref|NP_172223.1| protein phosphatase 2C (PP2C), putative; protei... 55 5e-07
ref|NP_180499.1| protein phosphatase 2C (PP2C), putative; protei... 49 3e-05
>emb|CAB90633.1| protein phpsphatase 2C (PP2C) [Fagus sylvatica]
Length = 413
Score = 63.9 bits (154), Expect = 1e-09
Identities = 30/61 (49%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Frame = -3
Query: 541 VATMCKERLHEIAKE-VNFVPENVEWKNTLKQRFICRDDEVQGWKSYINETTCRCELWTP 365
VA C++RLH+I KE V V E +EWK+T+++ F D EVQ W+ I T CRC++ TP
Sbjct: 161 VAMRCRDRLHDIVKEEVESVTEGMEWKDTMEKSFDRMDKEVQEWRVPIKTTNCRCDVQTP 220
Query: 364 Q 362
Q
Sbjct: 221 Q 221
>emb|CAC09575.1| protein phosphatase 2C (PP2C) [Fagus sylvatica]
Length = 183
Score = 62.4 bits (150), Expect = 4e-09
Identities = 30/61 (49%), Positives = 40/61 (65%), Gaps = 1/61 (1%)
Frame = -3
Query: 541 VATMCKERLHEIAKE-VNFVPENVEWKNTLKQRFICRDDEVQGWKSYINETTCRCELWTP 365
VA C++RLH+I KE V V E +EWK+T+++ F D EVQ W I T CRC++ TP
Sbjct: 8 VAMRCRDRLHDIVKEEVESVTEGMEWKDTMEKSFDRMDKEVQEWLVPIKTTNCRCDVQTP 67
Query: 364 Q 362
Q
Sbjct: 68 Q 68
>gb|AAM61361.1| protein phosphatase 2C, putative [Arabidopsis thaliana]
Length = 442
Score = 55.5 bits (132), Expect = 5e-07
Identities = 25/59 (42%), Positives = 32/59 (53%)
Frame = -3
Query: 541 VATMCKERLHEIAKEVNFVPENVEWKNTLKQRFICRDDEVQGWKSYINETTCRCELWTP 365
VA CKERLHE+ +E + EWK +++ F D EV W + CRCEL TP
Sbjct: 169 VAARCKERLHELVQEEALSDKKEEWKKMMERSFTRMDKEVVRWGETVMSANCRCELQTP 227
>ref|NP_172223.1| protein phosphatase 2C (PP2C), putative; protein id: At1g07430.1,
supported by cDNA: 118185., supported by cDNA:
gi_17979174, supported by cDNA: gi_20259128 [Arabidopsis
thaliana] gi|25352229|pir||B86209 protein F22G5.22
[imported] - Arabidopsis thaliana
gi|8778547|gb|AAF79555.1|AC022464_13 F22G5.22
[Arabidopsis thaliana] gi|17979175|gb|AAL49783.1|
putative protein phosphatase 2C [Arabidopsis thaliana]
gi|20259129|gb|AAM14280.1| putative phosphatase 2C
[Arabidopsis thaliana]
Length = 442
Score = 55.5 bits (132), Expect = 5e-07
Identities = 25/59 (42%), Positives = 32/59 (53%)
Frame = -3
Query: 541 VATMCKERLHEIAKEVNFVPENVEWKNTLKQRFICRDDEVQGWKSYINETTCRCELWTP 365
VA CKERLHE+ +E + EWK +++ F D EV W + CRCEL TP
Sbjct: 169 VAARCKERLHELVQEEALSDKKEEWKKMMERSFTRMDKEVVRWGETVMSANCRCELQTP 227
>ref|NP_180499.1| protein phosphatase 2C (PP2C), putative; protein id: At2g29380.1
[Arabidopsis thaliana] gi|25352214|pir||F84695 probable
protein phosphatase 2C [imported] - Arabidopsis thaliana
gi|3980397|gb|AAC95200.1| putative protein phosphatase
2C [Arabidopsis thaliana]
Length = 362
Score = 49.3 bits (116), Expect = 3e-05
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 2/61 (3%)
Frame = -3
Query: 541 VATMCKERLHEIAKEV--NFVPENVEWKNTLKQRFICRDDEVQGWKSYINETTCRCELWT 368
VA C+ERLH++ +E + + + EWK T+++ F D EV W + C+C+L T
Sbjct: 124 VAARCRERLHKLVQEELSSDMEDEEEWKTTMERSFTRMDKEVVSWGDSVVTANCKCDLQT 183
Query: 367 P 365
P
Sbjct: 184 P 184
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 476,011,710
Number of Sequences: 1393205
Number of extensions: 10405248
Number of successful extensions: 26764
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 26143
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26750
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)