Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001249A_C01 KMC001249A_c01
(1118 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
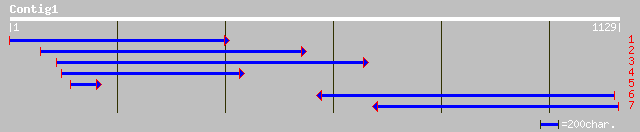
ref|NP_198440.1| nucleolar protein-like; protein id: At5g35910.1... 135 9e-31
gb|AAB03678.1| Rod1p 43 0.010
ref|NP_731502.1| CG14685-PA [Drosophila melanogaster] gi|1072644... 42 0.017
gb|AAN71089.1| AT19733p [Drosophila melanogaster] 42 0.017
ref|NP_731503.1| CG14685-PC [Drosophila melanogaster] gi|2317093... 42 0.017
>ref|NP_198440.1| nucleolar protein-like; protein id: At5g35910.1 [Arabidopsis
thaliana]
Length = 838
Score = 135 bits (341), Expect = 9e-31
Identities = 104/284 (36%), Positives = 150/284 (52%), Gaps = 23/284 (8%)
Frame = -3
Query: 1068 EQNKDANTGALSTLEGNG------------ATVQVLKKPAGAFGALLGNSASKRKHDPD- 928
E +D GA +T EG G A V++ KKP+ GALLGN+ASK+K D
Sbjct: 574 ESTRDLIMGAANTNEGRGLGSGLFGSAKVSAAVRISKKPSSGLGALLGNAASKKKSRTDE 633
Query: 927 KGMEEIKLEQIRSSVSLPFHSFLGSSEKSKPIVE-APSLASEMSDLHKPASDPVSTSTLD 751
K E++KLEQIRSSV+L FHSF SK E +P + + ++ ++ P S S D
Sbjct: 634 KVKEDVKLEQIRSSVNLSFHSFTEKVPDSKSTSETSPKVYGKPEEMS--STMPASVSKED 691
Query: 750 EIILL--DSDIAAEDLEQSNL--EDPIENRENRSTVSTSGTED-----EDVPMSLSELSS 598
+ L DS+ A+E + S E + + E + ++ ED PMSLSELS+
Sbjct: 692 GVKELKDDSEEASEIVGTSGRVSESKVSSSEMGDIILLENGDEKKVDAEDEPMSLSELST 751
Query: 597 NFQKCFQSNNHNTKTRQPQPTDPSALFQLKPFDYKAAMKFGENTARASSRTGDGHIEIEE 418
NFQKCF+S N + K ++ + ++PFDY+A AR + G+GH +
Sbjct: 752 NFQKCFKSMNKSKKAQK-----QTEFLNIEPFDYEA--------ARKEVKFGEGH----K 794
Query: 417 DSGGKKKRSHRGQGQSTDDLNKPLPQGRRRQAFPASGNRSATFR 286
GK++ + GQ + + QG+RRQAFPASGNRS +F+
Sbjct: 795 GRQGKREAA-AGQKKGSTQEQSEFGQGKRRQAFPASGNRSMSFK 837
>gb|AAB03678.1| Rod1p
Length = 837
Score = 42.7 bits (99), Expect = 0.010
Identities = 53/215 (24%), Positives = 83/215 (37%), Gaps = 28/215 (13%)
Frame = -3
Query: 831 VEAPSLASEMSDLHKPASDP---VSTSTLDEIILLD------------SDIAAEDLEQSN 697
+ +PSL + L + SD STS +++ L D SD+ EDL +
Sbjct: 600 LNSPSLTPSFAHLSRRNSDSRQTSSTSLKNDLELTDLSRVPSYDKAMKSDMIGEDLPPAY 659
Query: 696 LEDPIENREN------RSTVSTSGTEDEDVPMSLSELSSNFQKCFQSNNHNTKTRQPQPT 535
E+ + +EN R + + +P+ S SSN K S H + + P+
Sbjct: 660 PEEELGVQENKKIELERPQILHHKSTSSLLPLPGSSKSSNNLKRSSSRTHLSHSPLPRNN 719
Query: 534 DPSA--LFQLKPFDYKAAMKFGENTARASSRTGDGHIEIE-----EDSGGKKKRSHRGQG 376
S+ L QL + ++ + A S TG H + K SH +
Sbjct: 720 SGSSVSLQQLARNNTDSSFNLNLSFTSAKSSTGSRHFPFNMTTSFTSNSSSKNNSHFDKT 779
Query: 375 QSTDDLNKPLPQGRRRQAFPASGNRSATFR*YNEN 271
ST D NKP + A +RS++ R N N
Sbjct: 780 DSTSDANKPREEENYTSATHNRRSRSSSVRSNNSN 814
>ref|NP_731502.1| CG14685-PA [Drosophila melanogaster] gi|10726443|gb|AAF54547.2|
CG14685-PA [Drosophila melanogaster]
Length = 966
Score = 42.0 bits (97), Expect = 0.017
Identities = 22/85 (25%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Frame = -3
Query: 912 IKLEQIRSSVSLP-FHSFLGSSEKSKPIVEAPSLASEMSDLHKPASDPVSTSTLDEIILL 736
+K ++ +++S+ + L S++ P + + +E S L DP+ TLDE++L+
Sbjct: 372 LKSSEVEATLSVSHIENSLSDSQEENPPLSSTLAINESSVLDSTRVDPIKDLTLDELLLI 431
Query: 735 DSDIAAEDLEQSNLEDPIENRENRS 661
DS I+ E+L + +E++ S
Sbjct: 432 DSGISMEELSDIQMHTDLEDQRQLS 456
>gb|AAN71089.1| AT19733p [Drosophila melanogaster]
Length = 699
Score = 42.0 bits (97), Expect = 0.017
Identities = 22/85 (25%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Frame = -3
Query: 912 IKLEQIRSSVSLP-FHSFLGSSEKSKPIVEAPSLASEMSDLHKPASDPVSTSTLDEIILL 736
+K ++ +++S+ + L S++ P + + +E S L DP+ TLDE++L+
Sbjct: 194 LKSSEVEATLSVSHIENSLSDSQEENPPLSSTLAINESSVLDSTRVDPIKDLTLDELLLI 253
Query: 735 DSDIAAEDLEQSNLEDPIENRENRS 661
DS I+ E+L + +E++ S
Sbjct: 254 DSGISMEELSDIQMHTDLEDQRQLS 278
>ref|NP_731503.1| CG14685-PC [Drosophila melanogaster] gi|23170938|gb|AAN13485.1|
CG14685-PC [Drosophila melanogaster]
Length = 788
Score = 42.0 bits (97), Expect = 0.017
Identities = 22/85 (25%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Frame = -3
Query: 912 IKLEQIRSSVSLP-FHSFLGSSEKSKPIVEAPSLASEMSDLHKPASDPVSTSTLDEIILL 736
+K ++ +++S+ + L S++ P + + +E S L DP+ TLDE++L+
Sbjct: 194 LKSSEVEATLSVSHIENSLSDSQEENPPLSSTLAINESSVLDSTRVDPIKDLTLDELLLI 253
Query: 735 DSDIAAEDLEQSNLEDPIENRENRS 661
DS I+ E+L + +E++ S
Sbjct: 254 DSGISMEELSDIQMHTDLEDQRQLS 278
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 911,988,637
Number of Sequences: 1393205
Number of extensions: 20020862
Number of successful extensions: 52618
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 49089
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 52393
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 67811039634
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)