Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001247A_C01 KMC001247A_c01
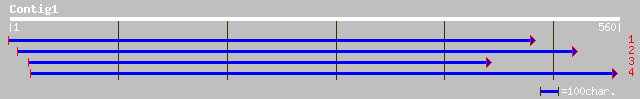
(560 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA92999.1| unnamed protein product [Oryza sativa (japonica ... 230 1e-59
pir||F96561 unknown protein [imported] - Arabidopsis thaliana gi... 223 1e-57
gb|AAN31930.1| unknown protein [Arabidopsis thaliana] 223 1e-57
ref|NP_175628.2| unknown protein; protein id: At1g52160.1, suppo... 223 1e-57
gb|AAM67367.1| unknown [Arabidopsis thaliana] 221 4e-57
>dbj|BAA92999.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 805
Score = 230 bits (586), Expect = 1e-59
Identities = 107/143 (74%), Positives = 128/143 (88%)
Frame = -3
Query: 555 AEERTNSVGKVIPGWKLVYSGDTRPCPELVEASQGATVLIHEATFEDALVDEALARNHST 376
A+E+ +S GK IPGWK+VYSGDTRPCP LV+AS+ ATVLIHEATFED++ DEA+ARNHST
Sbjct: 662 AKEKVSSAGKAIPGWKVVYSGDTRPCPALVDASRDATVLIHEATFEDSMKDEAIARNHST 721
Query: 375 TNEAIEMGNSANAYRIILTHFSQRYPKIPVLDETHMHKTCVAFDMMSINIADLPVLPKVL 196
T EAI +G SA AYRIILTHFSQRYPKIPV DE M KTC+AFD+MS+N+ADLPVLPKVL
Sbjct: 722 TKEAIAVGTSAGAYRIILTHFSQRYPKIPVFDEVDMQKTCIAFDLMSVNLADLPVLPKVL 781
Query: 195 PYLKLLFRNEMIVDESDDVVDAV 127
P+LKLLF++EM+VDESD++ +AV
Sbjct: 782 PHLKLLFKDEMVVDESDEIQEAV 804
>pir||F96561 unknown protein [imported] - Arabidopsis thaliana
gi|6850339|gb|AAF29402.1|AC022354_1 unknown protein
[Arabidopsis thaliana]
Length = 837
Score = 223 bits (568), Expect = 1e-57
Identities = 105/140 (75%), Positives = 126/140 (90%), Gaps = 1/140 (0%)
Frame = -3
Query: 555 AEERTNSVGKVIPGWKLVYSGDTRPCPELVEASQGATVLIHEATFEDALVDEALARNHST 376
A ER NSVG+ I GWK+VYSGD+RPCPE VEAS+ AT+LIHEATFEDAL++EALA+NHST
Sbjct: 688 AAERVNSVGEQILGWKMVYSGDSRPCPETVEASRDATILIHEATFEDALIEEALAKNHST 747
Query: 375 TNEAIEMGNSANAYRIILTHFSQRYPKIPVLDETHMHKTCVAFDMMSINIADLPVLPKVL 196
T EAI++G++AN YRI+LTHFSQRYPKIPV+DE+HMH TC+AFD+MSIN+ADL VLPKVL
Sbjct: 748 TKEAIDVGSAANVYRIVLTHFSQRYPKIPVIDESHMHNTCIAFDLMSINMADLHVLPKVL 807
Query: 195 PYLKLLFRNEMIVDE-SDDV 139
PY K LFR+EM+ DE +DDV
Sbjct: 808 PYFKTLFRDEMVEDEDADDV 827
>gb|AAN31930.1| unknown protein [Arabidopsis thaliana]
Length = 872
Score = 223 bits (568), Expect = 1e-57
Identities = 105/140 (75%), Positives = 126/140 (90%), Gaps = 1/140 (0%)
Frame = -3
Query: 555 AEERTNSVGKVIPGWKLVYSGDTRPCPELVEASQGATVLIHEATFEDALVDEALARNHST 376
A ER NSVG+ I GWK+VYSGD+RPCPE VEAS+ AT+LIHEATFEDAL++EALA+NHST
Sbjct: 723 AAERVNSVGEQILGWKMVYSGDSRPCPETVEASRDATILIHEATFEDALIEEALAKNHST 782
Query: 375 TNEAIEMGNSANAYRIILTHFSQRYPKIPVLDETHMHKTCVAFDMMSINIADLPVLPKVL 196
T EAI++G++AN YRI+LTHFSQRYPKIPV+DE+HMH TC+AFD+MSIN+ADL VLPKVL
Sbjct: 783 TKEAIDVGSAANVYRIVLTHFSQRYPKIPVIDESHMHNTCIAFDLMSINMADLHVLPKVL 842
Query: 195 PYLKLLFRNEMIVDE-SDDV 139
PY K LFR+EM+ DE +DDV
Sbjct: 843 PYFKTLFRDEMVEDEDADDV 862
>ref|NP_175628.2| unknown protein; protein id: At1g52160.1, supported by cDNA:
gi_17979120 [Arabidopsis thaliana]
gi|17979121|gb|AAL49818.1| unknown protein [Arabidopsis
thaliana] gi|21436181|gb|AAM51378.1| unknown protein
[Arabidopsis thaliana]
Length = 890
Score = 223 bits (568), Expect = 1e-57
Identities = 105/140 (75%), Positives = 126/140 (90%), Gaps = 1/140 (0%)
Frame = -3
Query: 555 AEERTNSVGKVIPGWKLVYSGDTRPCPELVEASQGATVLIHEATFEDALVDEALARNHST 376
A ER NSVG+ I GWK+VYSGD+RPCPE VEAS+ AT+LIHEATFEDAL++EALA+NHST
Sbjct: 741 AAERVNSVGEQILGWKMVYSGDSRPCPETVEASRDATILIHEATFEDALIEEALAKNHST 800
Query: 375 TNEAIEMGNSANAYRIILTHFSQRYPKIPVLDETHMHKTCVAFDMMSINIADLPVLPKVL 196
T EAI++G++AN YRI+LTHFSQRYPKIPV+DE+HMH TC+AFD+MSIN+ADL VLPKVL
Sbjct: 801 TKEAIDVGSAANVYRIVLTHFSQRYPKIPVIDESHMHNTCIAFDLMSINMADLHVLPKVL 860
Query: 195 PYLKLLFRNEMIVDE-SDDV 139
PY K LFR+EM+ DE +DDV
Sbjct: 861 PYFKTLFRDEMVEDEDADDV 880
>gb|AAM67367.1| unknown [Arabidopsis thaliana]
Length = 509
Score = 221 bits (564), Expect = 4e-57
Identities = 101/138 (73%), Positives = 122/138 (88%)
Frame = -3
Query: 555 AEERTNSVGKVIPGWKLVYSGDTRPCPELVEASQGATVLIHEATFEDALVDEALARNHST 376
A ER N G IPGWK+VYSGDTRPCPE+VEAS+GATVLIHEATFEDALV+EA+A+NHST
Sbjct: 352 AAERKNIAGDEIPGWKMVYSGDTRPCPEMVEASKGATVLIHEATFEDALVEEAVAKNHST 411
Query: 375 TNEAIEMGNSANAYRIILTHFSQRYPKIPVLDETHMHKTCVAFDMMSINIADLPVLPKVL 196
T EAI++G+SA YR +LTHFSQRYPKIPV+DE+HMH TC+AFDMMSIN+ADL VLPK+L
Sbjct: 412 TKEAIKVGSSAGVYRTVLTHFSQRYPKIPVIDESHMHNTCIAFDMMSINMADLHVLPKIL 471
Query: 195 PYLKLLFRNEMIVDESDD 142
PY K LFRN+++ +E ++
Sbjct: 472 PYFKTLFRNQVVEEEEEE 489
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 447,160,246
Number of Sequences: 1393205
Number of extensions: 8763549
Number of successful extensions: 23303
Number of sequences better than 10.0: 177
Number of HSP's better than 10.0 without gapping: 22806
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23271
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)