Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001242A_C01 KMC001242A_c01
(641 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
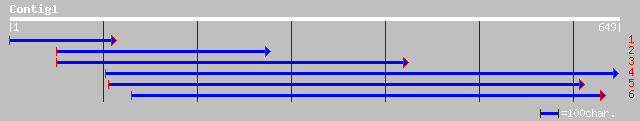
ref|NP_189087.1| hypothetical protein; protein id: At3g24440.1 [... 80 3e-14
ref|NP_194749.1| putative protein; protein id: At4g30200.1, supp... 64 2e-09
gb|AAO00859.1| putative protein [Arabidopsis thaliana] 64 2e-09
ref|NP_179477.1| unknown protein; protein id: At2g18870.1 [Arabi... 59 5e-08
ref|NP_179478.1| hypothetical protein; protein id: At2g18880.1 [... 58 9e-08
>ref|NP_189087.1| hypothetical protein; protein id: At3g24440.1 [Arabidopsis
thaliana] gi|9294095|dbj|BAB01947.1|
emb|CAB52464.1~gene_id:MXP5.1~similar to unknown protein
[Arabidopsis thaliana]
Length = 602
Score = 79.7 bits (195), Expect = 3e-14
Identities = 34/55 (61%), Positives = 47/55 (84%)
Frame = -3
Query: 636 FRLKLLTWFSLRSTEQERRLVNTFIQTLIDDPSTLAGPLVDSFSDIISSKRPRNG 472
FR++ LTWFS+ ST QE+ +V+TF+QTL DDP +LAG LVD+F+D++S+KRP NG
Sbjct: 543 FRVRFLTWFSMSSTAQEQSVVSTFVQTLEDDPGSLAGQLVDAFTDVVSTKRPNNG 597
>ref|NP_194749.1| putative protein; protein id: At4g30200.1, supported by cDNA:
gi_11177136 [Arabidopsis thaliana]
gi|7486763|pir||T14080 hypothetical protein F9N11.50 -
Arabidopsis thaliana gi|5730130|emb|CAB52464.1| putative
protein [Arabidopsis thaliana]
gi|7269920|emb|CAB81013.1| putative protein [Arabidopsis
thaliana] gi|11177137|dbj|BAB17836.1| nuclear
coiled-coil protein AT4g30200 [Arabidopsis thaliana]
Length = 714
Score = 63.9 bits (154), Expect = 2e-09
Identities = 34/70 (48%), Positives = 41/70 (58%), Gaps = 11/70 (15%)
Frame = -3
Query: 636 FRLKLLTWFSLRSTEQERRLVNTFIQTLIDDPSTLAGPLVDSFSDIISSKRPR------- 478
FR K LTW+SLR+T QE R+V FI T IDDP LA L+D+F D +S KR
Sbjct: 642 FRQKFLTWYSLRATSQEIRVVKIFIDTFIDDPMALAEQLIDTFDDRVSIKRSAVGGSGAS 701
Query: 477 ----NGFCSK 460
+GFC K
Sbjct: 702 AVVPSGFCMK 711
>gb|AAO00859.1| putative protein [Arabidopsis thaliana]
Length = 685
Score = 63.9 bits (154), Expect = 2e-09
Identities = 34/70 (48%), Positives = 41/70 (58%), Gaps = 11/70 (15%)
Frame = -3
Query: 636 FRLKLLTWFSLRSTEQERRLVNTFIQTLIDDPSTLAGPLVDSFSDIISSKRPR------- 478
FR K LTW+SLR+T QE R+V FI T IDDP LA L+D+F D +S KR
Sbjct: 613 FRQKFLTWYSLRATSQEIRVVKIFIDTFIDDPMALAEQLIDTFDDRVSIKRSAVGGSGAS 672
Query: 477 ----NGFCSK 460
+GFC K
Sbjct: 673 AVVPSGFCMK 682
>ref|NP_179477.1| unknown protein; protein id: At2g18870.1 [Arabidopsis thaliana]
gi|7485789|pir||T01615 hypothetical protein At2g18870
[imported] - Arabidopsis thaliana
gi|3004550|gb|AAC09023.1| unknown protein [Arabidopsis
thaliana]
Length = 302
Score = 58.9 bits (141), Expect = 5e-08
Identities = 30/50 (60%), Positives = 33/50 (66%)
Frame = -3
Query: 636 FRLKLLTWFSLRSTEQERRLVNTFIQTLIDDPSTLAGPLVDSFSDIISSK 487
FR K LTWFSLR+T QER V TF+ DD LA LVD+FSD I SK
Sbjct: 75 FRQKFLTWFSLRATSQERNTVTTFMNAFNDDSMALAEQLVDTFSDCIWSK 124
>ref|NP_179478.1| hypothetical protein; protein id: At2g18880.1 [Arabidopsis
thaliana] gi|7485790|pir||T01616 hypothetical protein
At2g18880 [imported] - Arabidopsis thaliana
gi|3004551|gb|AAC09024.1| hypothetical protein
[Arabidopsis thaliana]
Length = 417
Score = 58.2 bits (139), Expect = 9e-08
Identities = 25/56 (44%), Positives = 37/56 (65%)
Frame = -3
Query: 639 EFRLKLLTWFSLRSTEQERRLVNTFIQTLIDDPSTLAGPLVDSFSDIISSKRPRNG 472
+FR K LTW+ L++T++E+ +V F+ T DD LA L+D+FSD I+ K P G
Sbjct: 347 DFRKKFLTWYCLKATDKEKHVVEIFVDTFKDDKEALAKQLIDTFSDCITRKHPEIG 402
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 513,459,144
Number of Sequences: 1393205
Number of extensions: 10191765
Number of successful extensions: 22838
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 22234
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22834
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27007650415
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)