Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
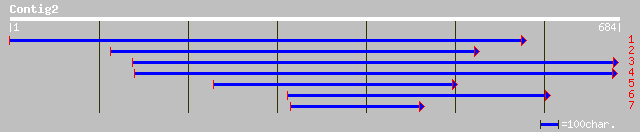
Query= KMC001210A_C02 KMC001210A_c02
(679 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_566784.1| expressed protein; protein id: At3g25910.1, sup... 35 0.94
ref|NP_700669.1| hypothetical protein [Plasmodium falciparum 3D7... 35 1.2
emb|CAA21941.1| serine /threonine protein kinase [Candida albicans] 34 2.1
sp|P50473|ARS_STRPU ARYLSULFATASE PRECURSOR (ARYL-SULFATE SULPHO... 33 4.7
pir||T31613 hypothetical protein Y50E8A.i - Caenorhabditis elegans 32 6.1
>ref|NP_566784.1| expressed protein; protein id: At3g25910.1, supported by cDNA:
gi_15983782 [Arabidopsis thaliana]
gi|9279596|dbj|BAB01054.1|
gb|AAF16548.1~gene_id:MPE11.8~similar to unknown protein
[Arabidopsis thaliana] gi|15983783|gb|AAL10488.1|
AT3g25910/MPE11_6 [Arabidopsis thaliana]
gi|23505907|gb|AAN28813.1| At3g25910/MPE11_6
[Arabidopsis thaliana]
Length = 372
Score = 35.0 bits (79), Expect = 0.94
Identities = 19/32 (59%), Positives = 22/32 (68%), Gaps = 1/32 (3%)
Frame = -2
Query: 678 GTASRDDANNESSDGG-SDTRRRRVRRRVTPD 586
GT+ RD+ NN+SSD S TRRRR RRR D
Sbjct: 333 GTSPRDEENNQSSDEQVSGTRRRRSRRRTVID 364
>ref|NP_700669.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23495061|gb|AAN35393.1|AE014832_15 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 1005
Score = 34.7 bits (78), Expect = 1.2
Identities = 18/51 (35%), Positives = 26/51 (50%)
Frame = -1
Query: 169 LMIYLYFFYLRFSYKYIYEEVFIFTPFSSTFHHVL*SFMNCLYFNSIFVFF 17
+ IY+YFF L Y YIY ++I+ FH V ++ +F F FF
Sbjct: 34 IYIYIYFFILSIIYIYIYIYIYIY------FHFVYYIYIYIFFFFFFFFFF 78
>emb|CAA21941.1| serine /threonine protein kinase [Candida albicans]
Length = 812
Score = 33.9 bits (76), Expect = 2.1
Identities = 16/54 (29%), Positives = 31/54 (56%)
Frame = +1
Query: 313 TQQKILFQAQLFTDLEKLQQRCSKNAK*TDYGIFNEHFSHSLATRQHKPKLIHN 474
T +K+L +A+ FT L++L R + + ++ I + H S L+ H K+++N
Sbjct: 117 TGRKVLSKAETFTHLQQLDTRNAAKNQLRNHRIPSNHLSSPLSAAPHSDKIVYN 170
>sp|P50473|ARS_STRPU ARYLSULFATASE PRECURSOR (ARYL-SULFATE SULPHOHYDROLASE) (ARS)
gi|283747|pir||A37362 arylsulfatase (EC 3.1.6.1)
precursor - sea urchin (Strongylocentrotus purpuratus)
gi|161441|gb|AAA30036.1| arylsulfatase
Length = 567
Score = 32.7 bits (73), Expect = 4.7
Identities = 21/78 (26%), Positives = 34/78 (42%)
Frame = +1
Query: 322 KILFQAQLFTDLEKLQQRCSKNAK*TDYGIFNEHFSHSLATRQHKPKLIHN*SEDPSRKS 501
K F+ Q + KL +RC DY + ++ +H P ++ + +DP
Sbjct: 433 KAHFKTQTDSSQMKLGERCDGGFPLDDYFLCSD--CEGDCVTEHNPPIMFDLEKDPGENY 490
Query: 502 PLSPSPLLHLLLENKNNI 555
PL P H+LL K+ I
Sbjct: 491 PLGPCGYEHVLLHVKDAI 508
>pir||T31613 hypothetical protein Y50E8A.i - Caenorhabditis elegans
Length = 836
Score = 32.3 bits (72), Expect = 6.1
Identities = 15/50 (30%), Positives = 28/50 (56%)
Frame = -1
Query: 166 MIYLYFFYLRFSYKYIYEEVFIFTPFSSTFHHVL*SFMNCLYFNSIFVFF 17
+ ++Y F+ +F +K+ + +FIF F F F+ L+F IF++F
Sbjct: 742 LFFIYLFFFKFFFKFFFLFLFIFFIFMFIF-----IFIFILFFIFIFIYF 786
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 574,288,072
Number of Sequences: 1393205
Number of extensions: 12487020
Number of successful extensions: 27584
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 26469
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27504
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29987172312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)