Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001190A_C01 KMC001190A_c01
(600 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
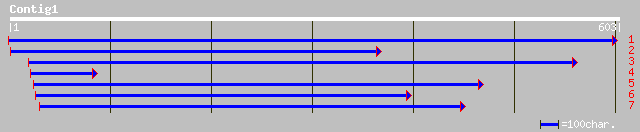
sp|O04893|AGLU_SPIOL ALPHA-GLUCOSIDASE PRECURSOR (MALTASE) gi|74... 86 3e-16
sp|O04931|AGLU_BETVU ALPHA-GLUCOSIDASE PRECURSOR (MALTASE) gi|74... 82 5e-15
gb|AAD05539.1| alpha-xylosidase precursor [Arabidopsis thaliana] 74 1e-12
ref|NP_177023.1| glycosyl hydrolase family 31 (alpha-xylosidase)... 74 1e-12
gb|AAB82656.1| alpha-glucosidase 1 [Arabidopsis thaliana] 67 1e-10
>sp|O04893|AGLU_SPIOL ALPHA-GLUCOSIDASE PRECURSOR (MALTASE) gi|7435426|pir||T09143
alpha-glucosidase (EC 3.2.1.20) - spinach
gi|2081627|dbj|BAA19924.1| alpha-glucosidase precoursor
[Spinacia oleracea]
Length = 903
Score = 86.3 bits (212), Expect = 3e-16
Identities = 42/116 (36%), Positives = 68/116 (58%), Gaps = 1/116 (0%)
Frame = -1
Query: 582 SYGQVYLDDGEALDMAGGKGQWTLVRFYSALQNNTVFVSSEVTNGRFALDQGWIIDKVTF 403
S+G+++LDDG + M +G+WT V+F +A T ++S+V +G FA+ Q W+IDKVT
Sbjct: 785 SFGELFLDDGVEVTMGVNRGKWTFVKFIAASAKQTCIITSDVVSGEFAVSQKWVIDKVTI 844
Query: 402 LGIPKNRRFGRMDLAVNG-TDSMRKTAVKTQIYTSSEFVIVEASKLSQLIGEQFKL 238
LG+ K + + T K+ +K+ EF++ E S L+ L+G +FKL
Sbjct: 845 LGLRKGTKINGYTVRTGAVTRKGDKSKLKSTPDRKGEFIVAEISGLNLLLGREFKL 900
>sp|O04931|AGLU_BETVU ALPHA-GLUCOSIDASE PRECURSOR (MALTASE) gi|7435427|pir||JC5463
alpha-glucosidase (EC 3.2.1.20) - sugar beet
gi|2190276|dbj|BAA20343.1| alpha-glucosidase [Beta
vulgaris]
Length = 913
Score = 82.0 bits (201), Expect = 5e-15
Identities = 40/119 (33%), Positives = 70/119 (58%), Gaps = 1/119 (0%)
Frame = -1
Query: 582 SYGQVYLDDGEALDMAGGKGQWTLVRFYSALQNNTVFVSSEVTNGRFALDQGWIIDKVTF 403
S G+++LD+G +D+ G G+WTLVRF++ N + +SSEV N +A+ Q W++DK+T
Sbjct: 789 STGELFLDNGIEMDIGGPGGKWTLVRFFAESGINNLTISSEVVNRGYAMSQRWVMDKITI 848
Query: 402 LGIPKNRRFGRMDLAVN-GTDSMRKTAVKTQIYTSSEFVIVEASKLSQLIGEQFKLEIQ 229
LG+ + + + + G ++ +T + F + S L QL+G+ FKLE++
Sbjct: 849 LGLKRRVKIKEYTVQKDAGAIKVKGLGRRTSSHNQGGFFVSVISDLRQLVGQAFKLELE 907
>gb|AAD05539.1| alpha-xylosidase precursor [Arabidopsis thaliana]
Length = 907
Score = 73.9 bits (180), Expect = 1e-12
Identities = 42/123 (34%), Positives = 74/123 (60%), Gaps = 10/123 (8%)
Frame = -1
Query: 576 GQVYLDDGEALDMAGGKGQWTLVRFYSALQNNTVFVSSEVTNGRFALDQGWIIDKVTFLG 397
G++YLD+ E +M G GQ T V FY+++ N T+ + S+V G+FAL +GW+I+KV+ LG
Sbjct: 781 GKLYLDEDELPEMKLGNGQSTYVDFYASVGNGTMKMWSQVKEGKFALSKGWVIEKVSVLG 840
Query: 396 IPKNRRFGRM-DLAVNGTDSMRKTAVKTQIYT---------SSEFVIVEASKLSQLIGEQ 247
+ R G++ ++ +NG+ +K V ++ +T ++ V+VE L L+G+
Sbjct: 841 L---RGAGQVSEIQINGSPMTKKIEVSSKEHTYVIGLEDEEENKSVMVEVRGLEMLVGKD 897
Query: 246 FKL 238
F +
Sbjct: 898 FNM 900
>ref|NP_177023.1| glycosyl hydrolase family 31 (alpha-xylosidase); protein id:
At1g68560.1, supported by cDNA: gi_15982750 [Arabidopsis
thaliana] gi|25404752|pir||H96709 hypothetical protein
F24J5.20 [imported] - Arabidopsis thaliana
gi|5002212|gb|AAD37363.1|AF144078_1 alpha-xylosidase
precursor [Arabidopsis thaliana]
gi|5734722|gb|AAD49987.1|AC008075_20 Identical to
gb|AF144078 alpha-xylosidase precursor from Arabidopsis
thaliana. ESTs gb|W43892, gb|N96165, gb|T46694,
gb|N37141, gb|R64965, gb|R90271, gb|AA651443,
gb|AA712305, gb|T04189 and gb|AA597852 come from this
gene gi|15982751|gb|AAL09716.1| At1g68560/F24J5_10
[Arabidopsis thaliana] gi|27363344|gb|AAO11591.1|
At1g68560/F24J5_10 [Arabidopsis thaliana]
Length = 915
Score = 73.9 bits (180), Expect = 1e-12
Identities = 42/123 (34%), Positives = 74/123 (60%), Gaps = 10/123 (8%)
Frame = -1
Query: 576 GQVYLDDGEALDMAGGKGQWTLVRFYSALQNNTVFVSSEVTNGRFALDQGWIIDKVTFLG 397
G++YLD+ E +M G GQ T V FY+++ N T+ + S+V G+FAL +GW+I+KV+ LG
Sbjct: 789 GKLYLDEDELPEMKLGNGQSTYVDFYASVGNGTMKMWSQVKEGKFALSKGWVIEKVSVLG 848
Query: 396 IPKNRRFGRM-DLAVNGTDSMRKTAVKTQIYT---------SSEFVIVEASKLSQLIGEQ 247
+ R G++ ++ +NG+ +K V ++ +T ++ V+VE L L+G+
Sbjct: 849 L---RGAGQVSEIQINGSPMTKKIEVSSKEHTYVIGLEDEEENKSVMVEVRGLEMLVGKD 905
Query: 246 FKL 238
F +
Sbjct: 906 FNM 908
>gb|AAB82656.1| alpha-glucosidase 1 [Arabidopsis thaliana]
Length = 902
Score = 67.4 bits (163), Expect = 1e-10
Identities = 42/121 (34%), Positives = 65/121 (53%), Gaps = 4/121 (3%)
Frame = -1
Query: 576 GQVYLDDGEALDMAGGKGQ--WTLVRFYSALQNNTVFVSSEVTNGRFALDQGWIIDKVTF 403
G+++LDDGE L M G G WTLV+F + +V + SEV N +A W I KVTF
Sbjct: 781 GELFLDDGENLRMGAGGGNRDWTLVKFRCYVTGKSVVLRSEVVNPEYASKMKWSIGKVTF 840
Query: 402 LGIPKNRRFGRMDLAVN-GTDSMRKTAVKT-QIYTSSEFVIVEASKLSQLIGEQFKLEIQ 229
+G ++ + S R + +KT F+ VE SKLS L+G++F++ ++
Sbjct: 841 VGFENVENVKTYEVRTSERLRSPRISLIKTVSDNDDPRFLSVEVSKLSLLVGKKFEMRLR 900
Query: 228 I 226
+
Sbjct: 901 L 901
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,227,983
Number of Sequences: 1393205
Number of extensions: 10346962
Number of successful extensions: 23397
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 22823
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23389
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)