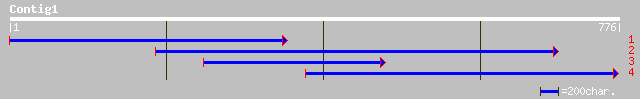
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001184A_C01 KMC001184A_c01
(776 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL91622.1| At2g23450/F26B6.10 [Arabidopsis thaliana] gi|2346... 96 8e-19
ref|NP_565552.1| putative protein kinase; protein id: At2g23450.... 96 8e-19
pir||T01134 probable protein kinase [imported] - Arabidopsis tha... 96 8e-19
gb|AAN52750.1| Putative protein kinase [Oryza sativa (japonica c... 78 2e-13
ref|XP_207272.1| similar to T cell receptor alpha [Mus musculus] 35 1.2
>gb|AAL91622.1| At2g23450/F26B6.10 [Arabidopsis thaliana]
gi|23463045|gb|AAN33192.1| At2g23450/F26B6.10
[Arabidopsis thaliana]
Length = 708
Score = 95.5 bits (236), Expect = 8e-19
Identities = 57/111 (51%), Positives = 72/111 (64%), Gaps = 1/111 (0%)
Frame = -1
Query: 776 FHSDMRPTMMEVAEELEHIRRSGWATMEETIGMASSVGSACSSPRNGSEKSVNSVKKVGQ 597
FHSDMRPTM EVA+ELE IR SGW ++ + S GS SS R GSE+SV K+
Sbjct: 605 FHSDMRPTMTEVADELEQIRLSGWIP---SMSLDSPAGSLRSSDR-GSERSV---KQSSI 657
Query: 596 ESEGLIVPPKDESYLQSME-VKDSSPVSVHDPWSSGHSSPSSNSLLENVVR 447
S +++P K L S+E + DSSP+SV DPW S SSPS+N+LL N+ R
Sbjct: 658 GSRRVVIPQKQPDCLASVEEISDSSPISVQDPWLSAQSSPSTNTLLGNIPR 708
>ref|NP_565552.1| putative protein kinase; protein id: At2g23450.1, supported by
cDNA: 19958., supported by cDNA: gi_19715596
[Arabidopsis thaliana] gi|20197000|gb|AAC23760.2|
putative protein kinase [Arabidopsis thaliana]
Length = 708
Score = 95.5 bits (236), Expect = 8e-19
Identities = 57/111 (51%), Positives = 72/111 (64%), Gaps = 1/111 (0%)
Frame = -1
Query: 776 FHSDMRPTMMEVAEELEHIRRSGWATMEETIGMASSVGSACSSPRNGSEKSVNSVKKVGQ 597
FHSDMRPTM EVA+ELE IR SGW ++ + S GS SS R GSE+SV K+
Sbjct: 605 FHSDMRPTMTEVADELEQIRLSGWIP---SMSLDSPAGSLRSSDR-GSERSV---KQSSI 657
Query: 596 ESEGLIVPPKDESYLQSME-VKDSSPVSVHDPWSSGHSSPSSNSLLENVVR 447
S +++P K L S+E + DSSP+SV DPW S SSPS+N+LL N+ R
Sbjct: 658 GSRRVVIPQKQPDCLASVEEISDSSPISVQDPWLSAQSSPSTNTLLGNIPR 708
>pir||T01134 probable protein kinase [imported] - Arabidopsis thaliana
Length = 694
Score = 95.5 bits (236), Expect = 8e-19
Identities = 57/111 (51%), Positives = 72/111 (64%), Gaps = 1/111 (0%)
Frame = -1
Query: 776 FHSDMRPTMMEVAEELEHIRRSGWATMEETIGMASSVGSACSSPRNGSEKSVNSVKKVGQ 597
FHSDMRPTM EVA+ELE IR SGW ++ + S GS SS R GSE+SV K+
Sbjct: 591 FHSDMRPTMTEVADELEQIRLSGWIP---SMSLDSPAGSLRSSDR-GSERSV---KQSSI 643
Query: 596 ESEGLIVPPKDESYLQSME-VKDSSPVSVHDPWSSGHSSPSSNSLLENVVR 447
S +++P K L S+E + DSSP+SV DPW S SSPS+N+LL N+ R
Sbjct: 644 GSRRVVIPQKQPDCLASVEEISDSSPISVQDPWLSAQSSPSTNTLLGNIPR 694
>gb|AAN52750.1| Putative protein kinase [Oryza sativa (japonica cultivar-group)]
Length = 704
Score = 77.8 bits (190), Expect = 2e-13
Identities = 50/108 (46%), Positives = 63/108 (58%), Gaps = 1/108 (0%)
Frame = -1
Query: 776 FHSDMRPTMMEVAEELEHIRRSGWATMEETIGMASSVGSACSS-PRNGSEKSVNSVKKVG 600
FHS+MRP+M EVA+ELE I+ SGWA + S+ S CSS P ++KS + K
Sbjct: 598 FHSEMRPSMAEVADELEQIQVSGWAPSTDDATFMSTTSSLCSSAPSRCTDKSWGTAKSKR 657
Query: 599 QESEGLIVPPKDESYLQSMEVKDSSPVSVHDPWSSGHSSPSSNSLLEN 456
Q + +V K E+ V D SPVSV + W S SSPSSNSLL N
Sbjct: 658 QAAANAVV--KQET--TKCAVAD-SPVSVQERWFSDRSSPSSNSLLRN 700
>ref|XP_207272.1| similar to T cell receptor alpha [Mus musculus]
Length = 278
Score = 35.0 bits (79), Expect = 1.2
Identities = 21/61 (34%), Positives = 30/61 (48%)
Frame = -1
Query: 644 RNGSEKSVNSVKKVGQESEGLIVPPKDESYLQSMEVKDSSPVSVHDPWSSGHSSPSSNSL 465
R G++ VNS +KV Q E LIVP E + S+ S S + W HS + +L
Sbjct: 157 RTGTQAGVNSQQKVQQSPESLIVP---EGAMTSLNCTFSDSASQYFAWYRQHSGKAPKAL 213
Query: 464 L 462
+
Sbjct: 214 M 214
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 651,069,888
Number of Sequences: 1393205
Number of extensions: 14195746
Number of successful extensions: 45749
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 43805
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 45707
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38375267554
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)