Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001170A_C01 KMC001170A_c01
(723 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
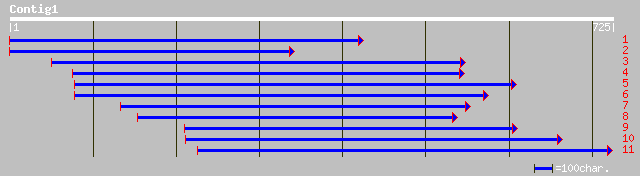
gb|AAK32887.1|AF367300_1 AT4g24840/F6I7_50 [Arabidopsis thaliana... 60 2e-08
ref|NP_567710.1| brefeldin A-sensitive Golgi protein - like; pro... 60 2e-08
gb|AAO37946.1| putative conserved oligomeric Golgi complex compo... 50 2e-05
pir||T06668 brefeldin A-sensitive Golgi protein LDLC homolog F6I... 45 0.001
emb|CAB93264.1| immunoglobulin heavy chain variable region [Homo... 33 4.0
>gb|AAK32887.1|AF367300_1 AT4g24840/F6I7_50 [Arabidopsis thaliana] gi|20147143|gb|AAM10288.1|
AT4g24840/F6I7_50 [Arabidopsis thaliana]
Length = 756
Score = 60.5 bits (145), Expect = 2e-08
Identities = 27/54 (50%), Positives = 39/54 (72%)
Frame = -3
Query: 634 KLCLLSFDNLPLAYCHQEYGRNLSALGVEAVNIASYCSLWQCVAPVDKKNSINL 473
K+C+ F ++ QEYGRN+SALG++ +I YCS WQCVAP D++NSI++
Sbjct: 709 KMCMQLFLDI------QEYGRNVSALGLKPADIPEYCSFWQCVAPADRQNSISV 756
Score = 45.4 bits (106), Expect = 8e-04
Identities = 20/30 (66%), Positives = 25/30 (82%)
Frame = -2
Query: 722 RRAGASSDISDNYVSDTDXICMQLFLDIQQ 633
RR GA+S +SD VS+TD +CMQLFLDIQ+
Sbjct: 691 RRGGAASGVSDQNVSETDKMCMQLFLDIQE 720
>ref|NP_567710.1| brefeldin A-sensitive Golgi protein - like; protein id:
At4g24840.1, supported by cDNA: gi_13605802, supported
by cDNA: gi_20147142 [Arabidopsis thaliana]
Length = 756
Score = 60.5 bits (145), Expect = 2e-08
Identities = 27/54 (50%), Positives = 39/54 (72%)
Frame = -3
Query: 634 KLCLLSFDNLPLAYCHQEYGRNLSALGVEAVNIASYCSLWQCVAPVDKKNSINL 473
K+C+ F ++ QEYGRN+SALG++ +I YCS WQCVAP D++NSI++
Sbjct: 709 KMCMQLFLDI------QEYGRNVSALGLKPADIPEYCSFWQCVAPADRQNSISV 756
Score = 45.4 bits (106), Expect = 8e-04
Identities = 20/30 (66%), Positives = 25/30 (82%)
Frame = -2
Query: 722 RRAGASSDISDNYVSDTDXICMQLFLDIQQ 633
RR GA+S +SD VS+TD +CMQLFLDIQ+
Sbjct: 691 RRGGAASGVSDQNVSETDKMCMQLFLDIQE 720
>gb|AAO37946.1| putative conserved oligomeric Golgi complex component [Oryza sativa
(japonica cultivar-group)]
Length = 754
Score = 50.4 bits (119), Expect = 2e-05
Identities = 22/30 (73%), Positives = 26/30 (86%)
Frame = -2
Query: 722 RRAGASSDISDNYVSDTDXICMQLFLDIQQ 633
RR GA++D SDN +SDTD ICMQLFLDIQ+
Sbjct: 689 RRVGANTDTSDNIISDTDKICMQLFLDIQE 718
Score = 48.9 bits (115), Expect = 7e-05
Identities = 23/52 (44%), Positives = 36/52 (69%)
Frame = -3
Query: 634 KLCLLSFDNLPLAYCHQEYGRNLSALGVEAVNIASYCSLWQCVAPVDKKNSI 479
K+C+ F ++ QEY RNL A+G++A I ++ +LWQCVAP DK+++I
Sbjct: 707 KICMQLFLDI------QEYARNLRAIGIDAREIETFRALWQCVAPRDKQDNI 752
>pir||T06668 brefeldin A-sensitive Golgi protein LDLC homolog F6I7.50 -
Arabidopsis thaliana gi|4678263|emb|CAB41124.1|
brefeldin A-sensitive Golgi protein-like [Arabidopsis
thaliana] gi|7269335|emb|CAB79394.1| brefeldin
A-sensitive Golgi protein-like [Arabidopsis thaliana]
Length = 724
Score = 44.7 bits (104), Expect = 0.001
Identities = 20/29 (68%), Positives = 24/29 (81%)
Frame = -2
Query: 722 RRAGASSDISDNYVSDTDXICMQLFLDIQ 636
RR GA+S +SD VS+TD +CMQLFLDIQ
Sbjct: 691 RRGGAASGVSDQNVSETDKMCMQLFLDIQ 719
>emb|CAB93264.1| immunoglobulin heavy chain variable region [Homo sapiens]
Length = 111
Score = 33.1 bits (74), Expect = 4.0
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Frame = +2
Query: 47 IYWDLQNKYQTTHENLFCN*LTYIKDVTKKQ-----TNIKGIEGNTYRCVRPIIKLL 202
IYWD +Y + ++ LT KD +K Q TNI ++ TY C R ++ +L
Sbjct: 47 IYWDDDKRYSPSLKSR----LTITKDTSKNQVVLTMTNIDPVDTATYYCARDVLDIL 99
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 573,687,701
Number of Sequences: 1393205
Number of extensions: 11668979
Number of successful extensions: 27587
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 26828
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27562
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33780557640
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)