Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001155A_C02 KMC001155A_c02
(629 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
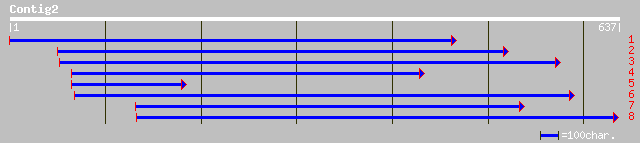
gb|AAK96502.1| At1g26300/F28B23_4 [Arabidopsis thaliana] gi|2265... 89 4e-17
ref|NP_173952.1| hypothetical protein; protein id: At1g26280.1 [... 89 4e-17
ref|NP_177066.1| hypothetical protein; protein id: At1g69030.1 [... 68 9e-11
gb|AAO51818.1| similar to Kaposi's sarcoma-associated herpesviru... 38 0.096
gb|AAN72216.1| Unknown protein [Arabidopsis thaliana] 37 0.21
>gb|AAK96502.1| At1g26300/F28B23_4 [Arabidopsis thaliana]
gi|22655044|gb|AAM98113.1| At1g26300/F28B23_4
[Arabidopsis thaliana]
Length = 316
Score = 89.4 bits (220), Expect = 4e-17
Identities = 47/74 (63%), Positives = 56/74 (75%), Gaps = 7/74 (9%)
Frame = -3
Query: 627 KAEATG------KNLK-SKSSNSSSEQDLDSFLLGDLEDSDGGPDDGEGSFDDDFDKIGN 469
KA ATG KN+ ++S +SSEQDLD+FLLGDLEDSD PDDG+GS +DDFDKIGN
Sbjct: 235 KAPATGETETVEKNVAIRRTSTASSEQDLDTFLLGDLEDSDEAPDDGDGSLEDDFDKIGN 294
Query: 468 SDVEDEKHVKKTSS 427
SDVEDEK K ++
Sbjct: 295 SDVEDEKQSSKATT 308
>ref|NP_173952.1| hypothetical protein; protein id: At1g26280.1 [Arabidopsis
thaliana] gi|25518376|pir||B86389 hypothetical protein
F28B23.6 - Arabidopsis thaliana
gi|12321170|gb|AAG50672.1|AC079829_5 hypothetical
protein [Arabidopsis thaliana]
Length = 98
Score = 89.4 bits (220), Expect = 4e-17
Identities = 47/74 (63%), Positives = 56/74 (75%), Gaps = 7/74 (9%)
Frame = -3
Query: 627 KAEATG------KNLK-SKSSNSSSEQDLDSFLLGDLEDSDGGPDDGEGSFDDDFDKIGN 469
KA ATG KN+ ++S +SSEQDLD+FLLGDLEDSD PDDG+GS +DDFDKIGN
Sbjct: 17 KAPATGETETVEKNVAIRRTSTASSEQDLDTFLLGDLEDSDEAPDDGDGSLEDDFDKIGN 76
Query: 468 SDVEDEKHVKKTSS 427
SDVEDEK K ++
Sbjct: 77 SDVEDEKQSSKATT 90
>ref|NP_177066.1| hypothetical protein; protein id: At1g69030.1 [Arabidopsis
thaliana] gi|25404796|pir||G96714 hypothetical protein
T6L1.21 [imported] - Arabidopsis thaliana
gi|12323210|gb|AAG51582.1|AC011665_3 hypothetical
protein [Arabidopsis thaliana]
Length = 320
Score = 68.2 bits (165), Expect = 9e-11
Identities = 35/54 (64%), Positives = 41/54 (75%), Gaps = 5/54 (9%)
Frame = -3
Query: 600 KSKSSNSSSEQDLDSFLLGDLEDSDGGPDDGEGS-----FDDDFDKIGNSDVED 454
K++ S +SSEQDLD+FLLGDLEDSD PDDG+G DDDFDKIGNS E+
Sbjct: 266 KNRISTASSEQDLDTFLLGDLEDSDEAPDDGDGDDGGSLGDDDFDKIGNSVSEN 319
>gb|AAO51818.1| similar to Kaposi's sarcoma-associated herpesvirus (KSHV) (Human
herpesvirus 8). ORF73 homolog [Dictyostelium discoideum]
Length = 3417
Score = 38.1 bits (87), Expect = 0.096
Identities = 24/89 (26%), Positives = 36/89 (39%)
Frame = +3
Query: 360 FKQQTSFSLESQTQVIAHTKLLQKMFSSHASHLQHQNCQSCQNHHQSCPHHHLDLHHCLQ 539
++QQ + Q Q H + Q H H QHQ Q Q H Q H H +Q
Sbjct: 1862 YQQQQHQQYQQQQQQQQHQQQHQHQHQQHQQHQQHQQHQQHQQHQQ-----HQQQHQLMQ 1916
Query: 540 GLQEEMNPSPAQRMNWKIYSSNSSLLLQP 626
Q+ + AQ+ K +++S + P
Sbjct: 1917 QRQQLETKNSAQQQQQKFQTNSSQVNNSP 1945
>gb|AAN72216.1| Unknown protein [Arabidopsis thaliana]
Length = 91
Score = 37.0 bits (84), Expect = 0.21
Identities = 24/66 (36%), Positives = 29/66 (43%)
Frame = +3
Query: 348 ISVSFKQQTSFSLESQTQVIAHTKLLQKMFSSHASHLQHQNCQSCQNHHQSCPHHHLDLH 527
+ V KQ+TS L S V + S H SHL HQ + N +S HHH H
Sbjct: 29 LPVLVKQETSLRLSSMPLV--------RDTSQHHSHLHHQLTPAAFNSAESDHHHHRSTH 80
Query: 528 HCLQGL 545
LQ L
Sbjct: 81 GTLQSL 86
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 482,107,497
Number of Sequences: 1393205
Number of extensions: 9665273
Number of successful extensions: 53420
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 40519
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49446
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)