Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001127A_C01 KMC001127A_c01
(438 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
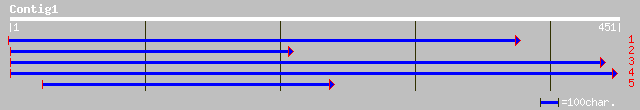
gb|EAA35202.1| predicted protein [Neurospora crassa] 39 0.015
ref|XP_085072.1| hypothetical protein XP_085072 [Homo sapiens] 37 0.073
ref|NP_645969.1| translation initiation factor IF-2 [Staphylococ... 37 0.073
ref|NP_371793.1| translation initiation factor IF-2 [Staphylococ... 37 0.073
gb|EAA18080.1| hypothetical protein [Plasmodium yoelii yoelii] 36 0.12
>gb|EAA35202.1| predicted protein [Neurospora crassa]
Length = 609
Score = 38.9 bits (89), Expect = 0.015
Identities = 19/82 (23%), Positives = 42/82 (51%)
Frame = +2
Query: 2 GSKSTLLELNRRQKHQAIQEKEAGQRQGS*QEERKGQEQKNRK*KQKKKSSKANKRNNRL 181
G K ++ ++QK Q Q+KE Q+Q Q++++ Q+Q+ ++ +Q+++ + +
Sbjct: 160 GQKQRQQQIEQQQKQQEQQQKEHEQQQQQQQQQQQQQQQQQQQQQQQQQQQNQQMQQMQQ 219
Query: 182 VNQTAQAINMAPHQQGLRVHNE 247
Q Q + QQ + H +
Sbjct: 220 AQQAQQIMQQTQQQQHQQQHQQ 241
>ref|XP_085072.1| hypothetical protein XP_085072 [Homo sapiens]
Length = 112
Score = 36.6 bits (83), Expect = 0.073
Identities = 22/68 (32%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Frame = +2
Query: 14 TLLELNRRQKHQAIQEKEAGQRQGS*QEERKGQ-EQKNRK*KQKKKS-SKANKRNNRLVN 187
T L+ R+K + +E++ G++ +E RK + E+K RK K++KK K K+ + V
Sbjct: 4 TFLKYCERRKKERKRERKEGRKGKEKRERRKKEKEEKERKKKERKKERKKERKKERKTVT 63
Query: 188 QTAQAINM 211
+T +A N+
Sbjct: 64 ETLKAENV 71
>ref|NP_645969.1| translation initiation factor IF-2 [Staphylococcus aureus subsp.
aureus MW2] gi|23821722|sp|Q8NWZ1|IF2_STAAW Translation
initiation factor IF-2 gi|21204320|dbj|BAB95017.1|
translation initiation factor IF-2 [Staphylococcus
aureus subsp. aureus MW2]
Length = 705
Score = 36.6 bits (83), Expect = 0.073
Identities = 21/69 (30%), Positives = 36/69 (51%), Gaps = 1/69 (1%)
Frame = +2
Query: 23 ELNRRQKHQAIQEKEAGQRQGS*QEERKGQEQKNRK*KQK-KKSSKANKRNNRLVNQTAQ 199
+ + QK+ Q + ++ + Q + KGQ++ N+K +Q+ K +K NK+NNR N
Sbjct: 49 KFKKEQKNDNKQSTQNNHQKSNNQNQNKGQQKDNKKNQQQNNKGNKGNKKNNR--NNKKN 106
Query: 200 AINMAPHQQ 226
N P Q
Sbjct: 107 NKNNKPQNQ 115
>ref|NP_371793.1| translation initiation factor IF-2 [Staphylococcus aureus subsp.
aureus Mu50] gi|15926852|ref|NP_374385.1| translation
initiation factor IF-2 [Staphylococcus aureus subsp.
aureus N315] gi|23821734|sp|Q99UK3|IF2_STAAM Translation
initiation factor IF-2 gi|25299472|pir||H89900
translation initiation factor IF-2 [imported] -
Staphylococcus aureus (strain N315)
gi|13701069|dbj|BAB42364.1| translation initiation
factor IF-2 [Staphylococcus aureus subsp. aureus N315]
gi|14247039|dbj|BAB57431.1| translation initiation
factor IF-2 [Staphylococcus aureus subsp. aureus Mu50]
Length = 705
Score = 36.6 bits (83), Expect = 0.073
Identities = 21/69 (30%), Positives = 36/69 (51%), Gaps = 1/69 (1%)
Frame = +2
Query: 23 ELNRRQKHQAIQEKEAGQRQGS*QEERKGQEQKNRK*KQK-KKSSKANKRNNRLVNQTAQ 199
+ + QK+ Q + ++ + Q + KGQ++ N+K +Q+ K +K NK+NNR N
Sbjct: 49 KFKKEQKNDNKQSTQNNHQKSNNQNQNKGQQKDNKKNQQQNNKGNKGNKKNNR--NNKKN 106
Query: 200 AINMAPHQQ 226
N P Q
Sbjct: 107 NKNNKPQNQ 115
>gb|EAA18080.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 357
Score = 35.8 bits (81), Expect = 0.12
Identities = 21/54 (38%), Positives = 32/54 (58%)
Frame = +3
Query: 42 STKLYRKKKQGSAKDPNKRNARAKNRRIESKNKKKRAAKQIKEITD**IKQHRQ 203
S K Y KKK S+ D R++R+K+ E KN+KK+ K+ E +D K+H +
Sbjct: 252 SDKSYYKKKSYSSSDEYSRSSRSKSYEYE-KNRKKKKKKKYNEDSDYKEKKHEK 304
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 348,541,101
Number of Sequences: 1393205
Number of extensions: 7098379
Number of successful extensions: 41084
Number of sequences better than 10.0: 421
Number of HSP's better than 10.0 without gapping: 32063
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38949
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 6722674608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)