Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
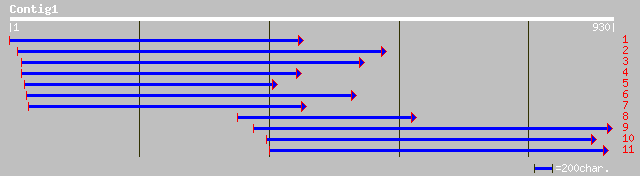
Query= KMC001114A_C01 KMC001114A_c01
(918 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_190012.1| CCR4-associated factor 1-like protein; protein ... 49 1e-04
ref|NP_704443.1| hypothetical protein [Plasmodium falciparum 3D7... 49 1e-04
gb|EAA20457.1| ccr4-not transcription complex, subunit 7 [Plasmo... 48 2e-04
ref|NP_197617.1| CCR4-associated factor-like protein; protein id... 47 3e-04
ref|NP_173044.1| BTG1 binding factor 1, putative; protein id: At... 45 0.002
>ref|NP_190012.1| CCR4-associated factor 1-like protein; protein id: At3g44260.1
[Arabidopsis thaliana] gi|11288396|pir||T49142
CCR4-associated factor 1-like protein - Arabidopsis
thaliana gi|7649377|emb|CAB88994.1| CCR4-associated
factor 1-like protein [Arabidopsis thaliana]
gi|15292829|gb|AAK92783.1| putative CCR4-associated
factor 1 [Arabidopsis thaliana]
gi|21436313|gb|AAM51295.1| putative CCR4-associated
factor 1 [Arabidopsis thaliana]
Length = 280
Score = 48.9 bits (115), Expect = 1e-04
Identities = 24/45 (53%), Positives = 33/45 (73%)
Frame = -1
Query: 276 LGRSHQVGSDSLLILHAFKKIMEVYFGNKDDGLVKYDGVLHGLEV 142
+G+ HQ GSDSLL HAF+++ ++YF DG K+ GVL+GLEV
Sbjct: 237 VGKCHQAGSDSLLTWHAFQRMRDLYF--VQDGPEKHAGVLYGLEV 279
>ref|NP_704443.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23499182|emb|CAD51262.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1774
Score = 48.9 bits (115), Expect = 1e-04
Identities = 23/43 (53%), Positives = 30/43 (69%)
Frame = -1
Query: 276 LGRSHQVGSDSLLILHAFKKIMEVYFGNKDDGLVKYDGVLHGL 148
+GR HQ GSDSL+ F K+ME+YF NK D KY G+++GL
Sbjct: 216 IGRQHQAGSDSLVTCKTFFKLMEMYFDNKIDD-KKYSGIIYGL 257
>gb|EAA20457.1| ccr4-not transcription complex, subunit 7 [Plasmodium yoelii
yoelii]
Length = 675
Score = 47.8 bits (112), Expect = 2e-04
Identities = 22/43 (51%), Positives = 30/43 (69%)
Frame = -1
Query: 276 LGRSHQVGSDSLLILHAFKKIMEVYFGNKDDGLVKYDGVLHGL 148
+GR HQ GSDSL+ F K++E+YF NK D KY G+++GL
Sbjct: 216 IGRQHQAGSDSLVTCKTFFKLLELYFDNKIDD-KKYSGIIYGL 257
>ref|NP_197617.1| CCR4-associated factor-like protein; protein id: At5g22250.1,
supported by cDNA: gi_17381057 [Arabidopsis thaliana]
gi|9757805|dbj|BAB08323.1| CCR4-associated factor-like
protein [Arabidopsis thaliana]
gi|17381058|gb|AAL36341.1| putative CCR4-associated
factor [Arabidopsis thaliana] gi|25054979|gb|AAN71961.1|
putative CCR4-associated factor [Arabidopsis thaliana]
Length = 278
Score = 47.4 bits (111), Expect = 3e-04
Identities = 23/45 (51%), Positives = 33/45 (73%)
Frame = -1
Query: 276 LGRSHQVGSDSLLILHAFKKIMEVYFGNKDDGLVKYDGVLHGLEV 142
+G+ HQ GSDSLL AF+++ ++YF +DG K+ GVL+GLEV
Sbjct: 235 VGKCHQAGSDSLLTWQAFQRMRDLYF--VEDGAEKHAGVLYGLEV 277
>ref|NP_173044.1| BTG1 binding factor 1, putative; protein id: At1g15920.1, supported
by cDNA: gi_19347886, supported by cDNA: gi_21281270
[Arabidopsis thaliana] gi|25363303|pir||F86293 T24D18.2
protein - Arabidopsis thaliana
gi|6587798|gb|AAF18489.1|AC010924_2 Similar to gi|Q60809
CCR4-associated factor 1 (CAF1) from Mus musculus. EST
gb|Z26822 comes from this gene. [Arabidopsis thaliana]
gi|19347887|gb|AAL86000.1| putative BTG1 binding factor
1 [Arabidopsis thaliana] gi|21281271|gb|AAM45088.1|
putative BTG1 binding factor 1 [Arabidopsis thaliana]
Length = 286
Score = 44.7 bits (104), Expect = 0.002
Identities = 25/51 (49%), Positives = 31/51 (60%)
Frame = -1
Query: 276 LGRSHQVGSDSLLILHAFKKIMEVYFGNKDDGLVKYDGVLHGLEV*KFLTG 124
+G SHQ GSDSLL L F K+ E +F L+KY G L GL+ + LTG
Sbjct: 236 VGISHQAGSDSLLTLRTFIKMKEFFF---TGSLLKYSGFLFGLDNPRLLTG 283
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 790,112,707
Number of Sequences: 1393205
Number of extensions: 17903670
Number of successful extensions: 40222
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 38221
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40176
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 50473155824
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)