Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001102A_C01 KMC001102A_c01
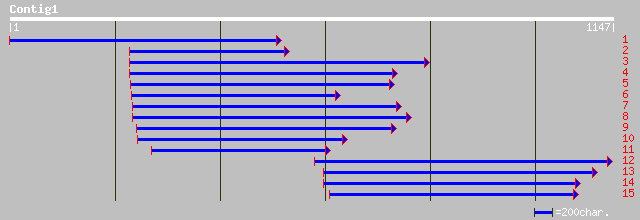
(1142 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_175737.1| hypothetical protein; protein id: At1g53300.1 [... 92 2e-17
dbj|BAB16491.1| P0665D10.16 [Oryza sativa (japonica cultivar-gro... 91 4e-17
dbj|BAB39114.2| P0686E09.6 [Oryza sativa (japonica cultivar-group)] 80 5e-14
ref|NP_188113.1| hypothetical protein; protein id: At3g14950.1 [... 78 2e-13
dbj|BAA97058.1| emb|CAB68200.1~gene_id:K15M2.9~strong similarity... 78 2e-13
>ref|NP_175737.1| hypothetical protein; protein id: At1g53300.1 [Arabidopsis thaliana]
gi|7769858|gb|AAF69536.1|AC008007_11 F12M16.20
[Arabidopsis thaliana]
Length = 699
Score = 91.7 bits (226), Expect = 2e-17
Identities = 41/56 (73%), Positives = 51/56 (90%)
Frame = -2
Query: 1141 PSINFLQVDIQESPTIATAENVRVVPTFKIYKNGSRVKEIICPSRDILEHTVRHYS 974
PSI+FL+VDI + P+I AENVRVVPT KIYKNGSRVKEI+CPS+++LE++VRHYS
Sbjct: 643 PSIHFLKVDIDKCPSIGNAENVRVVPTVKIYKNGSRVKEIVCPSKEVLEYSVRHYS 698
>dbj|BAB16491.1| P0665D10.16 [Oryza sativa (japonica cultivar-group)]
gi|21327945|dbj|BAC00538.1| P0489G09.5 [Oryza sativa
(japonica cultivar-group)]
Length = 862
Score = 90.5 bits (223), Expect = 4e-17
Identities = 41/55 (74%), Positives = 49/55 (88%)
Frame = -2
Query: 1141 PSINFLQVDIQESPTIATAENVRVVPTFKIYKNGSRVKEIICPSRDILEHTVRHY 977
PSI+FL+VDI ESP +A AENVR VPTFKIYKNG+RVKE+ICPS +LE++VRHY
Sbjct: 806 PSISFLKVDISESPAVARAENVRTVPTFKIYKNGTRVKEMICPSLQLLEYSVRHY 860
>dbj|BAB39114.2| P0686E09.6 [Oryza sativa (japonica cultivar-group)]
Length = 707
Score = 80.5 bits (197), Expect = 5e-14
Identities = 36/58 (62%), Positives = 50/58 (86%), Gaps = 2/58 (3%)
Frame = -2
Query: 1141 PSINFL--QVDIQESPTIATAENVRVVPTFKIYKNGSRVKEIICPSRDILEHTVRHYS 974
PS+NFL QV++++SP +A AENVR+VPTFKIYK+G +VKE+ICPS +L ++VRHY+
Sbjct: 646 PSLNFLKVQVNVEDSPMVAKAENVRIVPTFKIYKDGVKVKEMICPSLHVLRYSVRHYA 703
>ref|NP_188113.1| hypothetical protein; protein id: At3g14950.1 [Arabidopsis thaliana]
Length = 721
Score = 78.2 bits (191), Expect = 2e-13
Identities = 35/55 (63%), Positives = 45/55 (81%)
Frame = -2
Query: 1141 PSINFLQVDIQESPTIATAENVRVVPTFKIYKNGSRVKEIICPSRDILEHTVRHY 977
PS++FL+V+I + P + AE VRVVPTFKIYK G R+KEI+CPS++ LE TVRHY
Sbjct: 665 PSLHFLKVEIVKCPEVGNAERVRVVPTFKIYKLGIRMKEIVCPSKEALEKTVRHY 719
>dbj|BAA97058.1| emb|CAB68200.1~gene_id:K15M2.9~strong similarity to unknown protein
[Arabidopsis thaliana]
Length = 705
Score = 78.2 bits (191), Expect = 2e-13
Identities = 35/55 (63%), Positives = 45/55 (81%)
Frame = -2
Query: 1141 PSINFLQVDIQESPTIATAENVRVVPTFKIYKNGSRVKEIICPSRDILEHTVRHY 977
PS++FL+V+I + P + AE VRVVPTFKIYK G R+KEI+CPS++ LE TVRHY
Sbjct: 649 PSLHFLKVEIVKCPEVGNAERVRVVPTFKIYKLGIRMKEIVCPSKEALEKTVRHY 703
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,007,958,767
Number of Sequences: 1393205
Number of extensions: 24021124
Number of successful extensions: 75320
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 65783
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 73862
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 70007348610
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)