Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
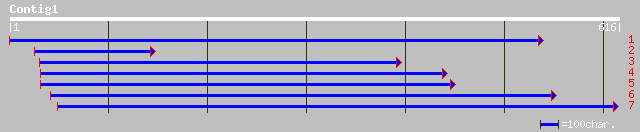
Query= KMC001070A_C01 KMC001070A_c01
(612 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_567467.1| F-box protein family, AtFBL4; protein id: At4g1... 139 2e-32
gb|AAM60829.1| F-box protein family, AtFBL4 [Arabidopsis thaliana] 139 2e-32
pir||C71419 hypothetical protein - Arabidopsis thaliana gi|22449... 139 2e-32
gb|AAK91884.1|AC091665_10 Putative leucine-rich repeats containi... 54 2e-06
ref|NP_197917.1| leucine-rich repeats containing protein; protei... 51 1e-05
>ref|NP_567467.1| F-box protein family, AtFBL4; protein id: At4g15475.1, supported by
cDNA: 100411., supported by cDNA: gi_13430831, supported
by cDNA: gi_13605654, supported by cDNA: gi_15810598
[Arabidopsis thaliana]
gi|13430832|gb|AAK26038.1|AF360328_1 putative F-box
protein family, AtFBL4 [Arabidopsis thaliana]
gi|13605655|gb|AAK32821.1|AF361808_1 AT4g15470/dl3775w
[Arabidopsis thaliana] gi|15810599|gb|AAL07187.1|
putative F-box protein family protein FBL4 [Arabidopsis
thaliana]
Length = 610
Score = 139 bits (351), Expect = 2e-32
Identities = 63/85 (74%), Positives = 75/85 (88%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 433
CP+++++V+SHC ITD GL HLV+ C +LE+CHMVYC GITSAGVATVVSSC +IKKVL
Sbjct: 526 CPMLKDLVLSHCHHITDNGLNHLVQKCKLLETCHMVYCPGITSAGVATVVSSCPHIKKVL 585
Query: 432 VEKWKVTPRTKRRAGSVISYLCVDL 358
+EKWKVT RT RRAGSVISYLC+DL
Sbjct: 586 IEKWKVTERTTRRAGSVISYLCMDL 610
Score = 52.0 bits (123), Expect = 6e-06
Identities = 20/63 (31%), Positives = 37/63 (57%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 433
CP ++E+ + +C++I + L + K C LE H+V CSGI + ++ C N+KK+
Sbjct: 371 CPRLKELALLYCQRIGNSALQEIGKGCKSLEILHLVDCSGIGDIAMCSIAKGCRNLKKLH 430
Query: 432 VEK 424
+ +
Sbjct: 431 IRR 433
Score = 42.7 bits (99), Expect = 0.004
Identities = 30/108 (27%), Positives = 51/108 (46%), Gaps = 7/108 (6%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSC-LNIKKV 436
C ++ + + C + D GLA + K C LE ++ +C G+T GV +V C ++K +
Sbjct: 165 CTSLKSLDLQGC-YVGDQGLAAVGKFCKQLEELNLRFCEGLTDVGVIDLVVGCSKSLKSI 223
Query: 435 -LVEKWKVTPRTKRRAGS-----VISYLCVDL*TSFGIYAYPQSCYRL 310
+ K+T + GS + YL + G+ A Q C+RL
Sbjct: 224 GVAASAKITDLSLEAVGSHCKLLEVLYLDSEYIHDKGLIAVAQGCHRL 271
Score = 33.9 bits (76), Expect = 1.7
Identities = 11/45 (24%), Positives = 25/45 (55%)
Frame = -1
Query: 570 ITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKV 436
+TD GL L +E+ +++C ++S G+ ++ C ++K +
Sbjct: 127 LTDTGLTALANGFPRIENLSLIWCPNVSSVGLCSLAQKCTSLKSL 171
Score = 32.0 bits (71), Expect = 6.5
Identities = 13/57 (22%), Positives = 28/57 (48%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIK 442
C + + I+ C I G+ + KSC L+ ++YC I ++ + + C +++
Sbjct: 345 CKELERVEINGCHNIGTRGIEAIGKSCPRLKELALLYCQRIGNSALQEIGKGCKSLE 401
>gb|AAM60829.1| F-box protein family, AtFBL4 [Arabidopsis thaliana]
Length = 610
Score = 139 bits (351), Expect = 2e-32
Identities = 63/85 (74%), Positives = 75/85 (88%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 433
CP+++++V+SHC ITD GL HLV+ C +LE+CHMVYC GITSAGVATVVSSC +IKKVL
Sbjct: 526 CPMLKDLVLSHCHHITDNGLNHLVQKCKLLETCHMVYCPGITSAGVATVVSSCPHIKKVL 585
Query: 432 VEKWKVTPRTKRRAGSVISYLCVDL 358
+EKWKVT RT RRAGSVISYLC+DL
Sbjct: 586 IEKWKVTERTTRRAGSVISYLCMDL 610
Score = 52.0 bits (123), Expect = 6e-06
Identities = 20/63 (31%), Positives = 37/63 (57%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 433
CP ++E+ + +C++I + L + K C LE H+V CSGI + ++ C N+KK+
Sbjct: 371 CPRLKELALLYCQRIGNSALQEIGKGCKSLEILHLVDCSGIGDIAMCSIAKGCRNLKKLH 430
Query: 432 VEK 424
+ +
Sbjct: 431 IRR 433
Score = 39.7 bits (91), Expect = 0.031
Identities = 29/108 (26%), Positives = 49/108 (44%), Gaps = 7/108 (6%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSC-LNIKKV 436
C ++ + + C + D GLA + K C LE ++ +C G+T GV + C ++K +
Sbjct: 165 CTSLKSLDLQGC-YVGDQGLAAVGKFCKQLEELNLRFCEGLTDVGVIDLAVGCSKSLKSI 223
Query: 435 -LVEKWKVTPRTKRRAGS-----VISYLCVDL*TSFGIYAYPQSCYRL 310
+ K+T + GS + YL + G+ A Q C RL
Sbjct: 224 GVAASAKITDLSLEAVGSHCKLLEVLYLDSEYIHDKGLIAVAQGCNRL 271
Score = 33.5 bits (75), Expect = 2.2
Identities = 11/45 (24%), Positives = 25/45 (55%)
Frame = -1
Query: 570 ITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKV 436
+TD GL L +E+ +++C ++S G+ ++ C ++K +
Sbjct: 127 LTDTGLTALADGFPRIENLSLIWCPNVSSVGLCSLAQKCTSLKSL 171
Score = 32.0 bits (71), Expect = 6.5
Identities = 13/57 (22%), Positives = 28/57 (48%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIK 442
C + + I+ C I G+ + KSC L+ ++YC I ++ + + C +++
Sbjct: 345 CKELERVEINGCHNIGTRGIEAIGKSCPRLKELALLYCQRIGNSALQEIGKGCKSLE 401
>pir||C71419 hypothetical protein - Arabidopsis thaliana
gi|2244904|emb|CAB10325.1| hypothetical protein
[Arabidopsis thaliana] gi|7268294|emb|CAB78589.1|
hypothetical protein [Arabidopsis thaliana]
Length = 712
Score = 139 bits (351), Expect = 2e-32
Identities = 63/85 (74%), Positives = 75/85 (88%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 433
CP+++++V+SHC ITD GL HLV+ C +LE+CHMVYC GITSAGVATVVSSC +IKKVL
Sbjct: 628 CPMLKDLVLSHCHHITDNGLNHLVQKCKLLETCHMVYCPGITSAGVATVVSSCPHIKKVL 687
Query: 432 VEKWKVTPRTKRRAGSVISYLCVDL 358
+EKWKVT RT RRAGSVISYLC+DL
Sbjct: 688 IEKWKVTERTTRRAGSVISYLCMDL 712
Score = 42.7 bits (99), Expect = 0.004
Identities = 30/108 (27%), Positives = 51/108 (46%), Gaps = 7/108 (6%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSC-LNIKKV 436
C ++ + + C + D GLA + K C LE ++ +C G+T GV +V C ++K +
Sbjct: 371 CTSLKSLDLQGC-YVGDQGLAAVGKFCKQLEELNLRFCEGLTDVGVIDLVVGCSKSLKSI 429
Query: 435 -LVEKWKVTPRTKRRAGS-----VISYLCVDL*TSFGIYAYPQSCYRL 310
+ K+T + GS + YL + G+ A Q C+RL
Sbjct: 430 GVAASAKITDLSLEAVGSHCKLLEVLYLDSEYIHDKGLIAVAQGCHRL 477
Score = 33.9 bits (76), Expect = 1.7
Identities = 11/45 (24%), Positives = 25/45 (55%)
Frame = -1
Query: 570 ITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKV 436
+TD GL L +E+ +++C ++S G+ ++ C ++K +
Sbjct: 333 LTDTGLTALANGFPRIENLSLIWCPNVSSVGLCSLAQKCTSLKSL 377
>gb|AAK91884.1|AC091665_10 Putative leucine-rich repeats containing protein [Oryza sativa]
Length = 628
Score = 53.5 bits (127), Expect = 2e-06
Identities = 22/63 (34%), Positives = 39/63 (60%)
Frame = -1
Query: 603 IREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVLVEK 424
++ +++ C I+DVGL + + C LE+C + CS +T AGVA + +++++VEK
Sbjct: 546 LKHLMMLRCDAISDVGLEDIARGCLQLEACGVFRCSQVTPAGVAALAGGSSRLQRIIVEK 605
Query: 423 WKV 415
KV
Sbjct: 606 CKV 608
Score = 32.0 bits (71), Expect = 6.5
Identities = 13/44 (29%), Positives = 24/44 (54%)
Frame = -1
Query: 567 TDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKV 436
TD GL HL++ C LE + + I+ G+ + + C N++ +
Sbjct: 141 TDAGLLHLIEGCKGLEKLTLNWFLHISEKGLVGIANRCRNLQSL 184
>ref|NP_197917.1| leucine-rich repeats containing protein; protein id: At5g25350.1
[Arabidopsis thaliana]
Length = 623
Score = 50.8 bits (120), Expect = 1e-05
Identities = 23/85 (27%), Positives = 46/85 (54%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 433
CP +R + + + ++D+GL+ + +SC M+E + C GIT +G+ + +C+N+ +
Sbjct: 166 CPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGITDSGLVAIAENCVNLSDLT 225
Query: 432 VEKWKVTPRTKRRAGSVISYLCVDL 358
++ RA I+ CV+L
Sbjct: 226 IDSCSGVGNEGLRA---IARRCVNL 247
Score = 48.1 bits (113), Expect = 9e-05
Identities = 18/61 (29%), Positives = 35/61 (56%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVL 433
CP+I ++ +S C ITD GL + ++C L + CSG+ + G+ + C+N++ +
Sbjct: 192 CPMIEKLDLSRCPGITDSGLVAIAENCVNLSDLTIDSCSGVGNEGLRAIARRCVNLRSIS 251
Query: 432 V 430
+
Sbjct: 252 I 252
Score = 33.5 bits (75), Expect = 2.2
Identities = 17/52 (32%), Positives = 25/52 (47%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSS 457
C +R + I C D LA L K C L+ + +G+T AGV ++ S
Sbjct: 431 CSSLRSLSIRCCPGFGDASLAFLGKFCHQLQDVELCGLNGVTDAGVRELLQS 482
Score = 33.1 bits (74), Expect = 2.9
Identities = 15/53 (28%), Positives = 26/53 (48%)
Frame = -1
Query: 612 CPLIREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSC 454
CP ++ + ++ C ++ GL L KS LES + C I G+ + +C
Sbjct: 350 CPDLKHVSLNKCLLVSGKGLVALAKSALSLESLKLEECHRINQFGLMGFLMNC 402
Score = 33.1 bits (74), Expect = 2.9
Identities = 14/60 (23%), Positives = 32/60 (53%)
Frame = -1
Query: 603 IREIVISHCRQITDVGLAHLVKSCTMLESCHMVYCSGITSAGVATVVSSCLNIKKVLVEK 424
++ + + CR +TDVGL + C L+ + C ++ G+ + S L+++ + +E+
Sbjct: 327 LKSLSVMSCRGMTDVGLEAVGNGCPDLKHVSLNKCLLVSGKGLVALAKSALSLESLKLEE 386
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 504,126,439
Number of Sequences: 1393205
Number of extensions: 10600017
Number of successful extensions: 19638
Number of sequences better than 10.0: 120
Number of HSP's better than 10.0 without gapping: 18836
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19599
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24568846532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)