Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001069A_C01 KMC001069A_c01
(522 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
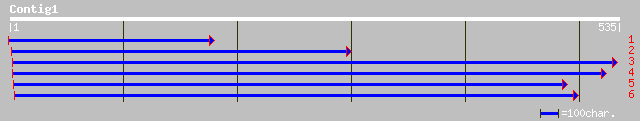
ref|NP_197170.1| putative protein; protein id: At5g16680.1 [Arab... 40 0.013
emb|CAC15053.1| envelope protein [Human immunodeficiency virus 1] 27 4.8
ref|XP_213225.1| similar to hypothetical protein XP_148064 [Mus ... 32 5.8
ref|NP_700792.1| hypothetical protein [Plasmodium falciparum 3D7... 32 5.8
ref|NP_010437.1| Epsin N-Terminal homology-containing protein; E... 31 7.6
>ref|NP_197170.1| putative protein; protein id: At5g16680.1 [Arabidopsis thaliana]
gi|11357809|pir||T51500 hypothetical protein F5E19_20 -
Arabidopsis thaliana gi|9755720|emb|CAC01832.1| putative
protein [Arabidopsis thaliana]
Length = 1280
Score = 40.4 bits (93), Expect = 0.013
Identities = 24/63 (38%), Positives = 33/63 (52%)
Frame = -2
Query: 521 AGTADKKNNQEKTPDLLEDEKKNDTDSVAASLSLSLSFPSSDKEQIRPVSESDQHVNAPL 342
+ + ++ NN DE++ D VAASLSLSLSFP +E ++VN PL
Sbjct: 1222 SNSGEQSNNSMNKEKQKADEEEEDDAEVAASLSLSLSFPG---------TEERKNVNTPL 1272
Query: 341 LLF 333
LF
Sbjct: 1273 FLF 1275
>emb|CAC15053.1| envelope protein [Human immunodeficiency virus 1]
Length = 840
Score = 27.3 bits (59), Expect(2) = 4.8
Identities = 12/47 (25%), Positives = 29/47 (61%)
Frame = +1
Query: 367 DSETGLICSLSEEGKDRDKERDAATLSVSFFFSSSNKSGVFSWLFFL 507
D+ TGLI +L E+ +++ ++ + L++ + S N + +WL+++
Sbjct: 620 DNYTGLIYTLIEDAQNQQEKNEQDLLALDKWASLWNWFDISNWLWYI 666
Score = 23.1 bits (48), Expect(2) = 4.8
Identities = 10/26 (38%), Positives = 15/26 (57%)
Frame = +3
Query: 237 LDDYIHDEQDRGIWNKMMNMIRNTRL 314
L+ Y+ D+Q GIW +I TR+
Sbjct: 567 LERYLRDQQLLGIWGCSGKLICTTRV 592
>ref|XP_213225.1| similar to hypothetical protein XP_148064 [Mus musculus] [Rattus
norvegicus]
Length = 172
Score = 31.6 bits (70), Expect = 5.8
Identities = 16/52 (30%), Positives = 30/52 (56%)
Frame = -2
Query: 518 GTADKKNNQEKTPDLLEDEKKNDTDSVAASLSLSLSFPSSDKEQIRPVSESD 363
G +D+ + E + +ED++++D D S++LS + PS + P+ ESD
Sbjct: 118 GDSDEDEDSEDSEMNMEDDEEDDDDLEDESIALSPAKPSRRVRKPEPLDESD 169
>ref|NP_700792.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23495186|gb|AAN35516.1|AE014834_13 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 553
Score = 31.6 bits (70), Expect = 5.8
Identities = 16/53 (30%), Positives = 28/53 (52%)
Frame = -2
Query: 509 DKKNNQEKTPDLLEDEKKNDTDSVAASLSLSLSFPSSDKEQIRPVSESDQHVN 351
D+KN+ EK D D++KND + + A ++ + EQI +D+ +N
Sbjct: 265 DEKNDDEKNDDEKNDDEKNDDEKIDAE---QINDEQINAEQINDEQINDEQIN 314
>ref|NP_010437.1| Epsin N-Terminal homology-containing protein; Ent5p [Saccharomyces
cerevisiae] gi|2132475|pir||S57979 probable membrane
protein YDR153c - yeast (Saccharomyces cerevisiae)
gi|899402|emb|CAA90375.1| unknown [Saccharomyces
cerevisiae]
Length = 411
Score = 31.2 bits (69), Expect = 7.6
Identities = 19/53 (35%), Positives = 29/53 (53%), Gaps = 1/53 (1%)
Frame = -2
Query: 515 TADKKNNQEKTPDLLE-DEKKNDTDSVAASLSLSLSFPSSDKEQIRPVSESDQ 360
T K + K DLL+ D K+DTD+ AA+ SL F +++ RP + D+
Sbjct: 342 TTAPKTSNSKIDDLLDWDGPKSDTDTTAAA-QTSLPFAEKKQQKARPQATKDK 393
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 423,937,338
Number of Sequences: 1393205
Number of extensions: 8649755
Number of successful extensions: 26254
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 25307
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26242
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 16731298976
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)