Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
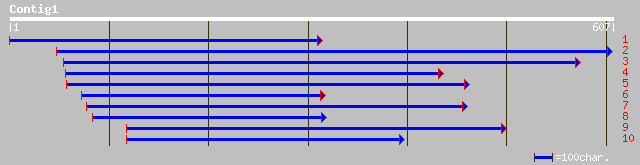
Query= KMC001047A_C01 KMC001047A_c01
(585 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_187461.1| MATE efflux family protein, putative; protein i... 141 7e-33
gb|AAL82672.1|AC092387_20 putative membrane protein [Oryza sativ... 118 6e-26
pir||D96551 hypothetical protein F11M15.20 [imported] - Arabidop... 109 3e-23
gb|AAO42040.1| unknown protein [Arabidopsis thaliana] gi|2897323... 95 6e-19
ref|NP_181367.1| hypothetical protein; protein id: At2g38330.1 [... 80 2e-14
>ref|NP_187461.1| MATE efflux family protein, putative; protein id: At3g08040.1,
supported by cDNA: gi_15912322, supported by cDNA:
gi_15982819, supported by cDNA: gi_19310968 [Arabidopsis
thaliana] gi|6648216|gb|AAF21214.1|AC013483_38 putative
integral membrane protein [Arabidopsis thaliana]
gi|12321554|gb|AAG50830.1|AC074395_4 MATE efflux family
protein, putative [Arabidopsis thaliana]
gi|15912323|gb|AAL08295.1| T8G24.8/T8G24.8 [Arabidopsis
thaliana] gi|15982820|gb|AAL09757.1| T8G24.8/T8G24.8
[Arabidopsis thaliana]
gi|19310969|gb|AAL86700.1|AF448231_1 ferric reductase
defective [Arabidopsis thaliana]
gi|22137274|gb|AAM91482.1| At3g08040/T8G24.8
[Arabidopsis thaliana]
Length = 526
Score = 141 bits (355), Expect = 7e-33
Identities = 66/102 (64%), Positives = 83/102 (80%)
Frame = -2
Query: 584 FSESAPVVHLIRIAVPFVAATQPIHALAFVFDGVNYGASDFAYSAYSMVMVSFATIASLF 405
FS+ V+HL+ I +PF+AATQPI++LAFV DGVN+GASDFAY+AYSMV V+ +IA++
Sbjct: 418 FSKDPAVIHLMAIGIPFIAATQPINSLAFVLDGVNFGASDFAYTAYSMVGVAAISIAAVI 477
Query: 404 LLYKSNGFIGIWIALTIYMTLRMFAGLFRMGTCSGPWRFLRG 279
+ K+NGFIGIWIALTIYM LR G+ RM T +GPWRFLRG
Sbjct: 478 YMAKTNGFIGIWIALTIYMALRAITGIARMATGTGPWRFLRG 519
>gb|AAL82672.1|AC092387_20 putative membrane protein [Oryza sativa (japonica cultivar-group)]
Length = 501
Score = 118 bits (295), Expect = 6e-26
Identities = 53/92 (57%), Positives = 71/92 (76%)
Frame = -2
Query: 584 FSESAPVVHLIRIAVPFVAATQPIHALAFVFDGVNYGASDFAYSAYSMVMVSFATIASLF 405
F++ V+H I + +PFV+ TQPI+ALAFVFDG+NYGASDF Y+AYSM++V+ +I +
Sbjct: 408 FTDDQDVLHHIYLGIPFVSLTQPINALAFVFDGINYGASDFGYAAYSMILVAIVSIIFIV 467
Query: 404 LLYKSNGFIGIWIALTIYMTLRMFAGLFRMGT 309
L NGF+GIWIALT+YM+LRM AG R+ T
Sbjct: 468 TLASYNGFVGIWIALTVYMSLRMLAGFLRINT 499
>pir||D96551 hypothetical protein F11M15.20 [imported] - Arabidopsis thaliana
gi|4836944|gb|AAD30646.1|AC006085_19 Hypothetical
protein [Arabidopsis thaliana]
Length = 501
Score = 109 bits (272), Expect = 3e-23
Identities = 52/89 (58%), Positives = 67/89 (74%)
Frame = -2
Query: 584 FSESAPVVHLIRIAVPFVAATQPIHALAFVFDGVNYGASDFAYSAYSMVMVSFATIASLF 405
F++ V+HLI I +PFVA TQPI+ALAFVFDGVN+GASDF Y+A S+VMV+ +I L
Sbjct: 413 FTKDDKVLHLISIGLPFVAGTQPINALAFVFDGVNFGASDFGYAAASLVMVAIVSILCLL 472
Query: 404 LLYKSNGFIGIWIALTIYMTLRMFAGLFR 318
L ++GFIG+W LTIYM+LR G +R
Sbjct: 473 FLSSTHGFIGLWFGLTIYMSLRAAVGFWR 501
>gb|AAO42040.1| unknown protein [Arabidopsis thaliana] gi|28973233|gb|AAO63941.1|
unknown protein [Arabidopsis thaliana]
Length = 521
Score = 95.1 bits (235), Expect = 6e-19
Identities = 48/102 (47%), Positives = 69/102 (67%), Gaps = 2/102 (1%)
Frame = -2
Query: 584 FSESAPVVHLIRIAVPFVAATQPIHALAFVFDGVNYGASDFAYSAYSMVMVSFATIASLF 405
F+ + V+ + FVA +QP++ALAFV DG+ YG SDF ++AYSMV+V F I+SLF
Sbjct: 414 FTTDSEVLKIALSGTLFVAGSQPVNALAFVLDGLYYGVSDFGFAAYSMVIVGF--ISSLF 471
Query: 404 LLYKSN--GFIGIWIALTIYMTLRMFAGLFRMGTCSGPWRFL 285
+L + G GIW L ++M LR+ AG +R+GT +GPW+ L
Sbjct: 472 MLVAAPTFGLAGIWTGLFLFMALRLVAGAWRLGTRTGPWKML 513
>ref|NP_181367.1| hypothetical protein; protein id: At2g38330.1 [Arabidopsis
thaliana] gi|7487160|pir||T02512 hypothetical protein
At2g38330 [imported] - Arabidopsis thaliana
gi|3395439|gb|AAC28771.1| hypothetical protein
[Arabidopsis thaliana]
Length = 539
Score = 79.7 bits (195), Expect = 2e-14
Identities = 42/92 (45%), Positives = 61/92 (65%), Gaps = 2/92 (2%)
Frame = -2
Query: 584 FSESAPVVHLIRIAVPFVAATQPIHALAFVFDGVNYGASDFAYSAYSMVMVSFATIASLF 405
F+ + V+ + FVA +QP++ALAFV DG+ YG SDF ++AYSMV+V F I+SLF
Sbjct: 414 FTTDSEVLKIALSGTLFVAGSQPVNALAFVLDGLYYGVSDFGFAAYSMVIVGF--ISSLF 471
Query: 404 LLYKSN--GFIGIWIALTIYMTLRMFAGLFRM 315
+L + G GIW L ++M LR+ AG +R+
Sbjct: 472 MLVAAPTFGLAGIWTGLFLFMALRLVAGAWRI 503
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 518,633,021
Number of Sequences: 1393205
Number of extensions: 11346179
Number of successful extensions: 22402
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 21811
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22390
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)