Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
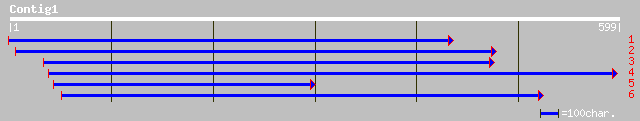
Query= KMC001045A_C01 KMC001045A_c01
(596 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM97031.1| serine carboxypeptidase 1 precursor-like protein ... 234 8e-61
dbj|BAB01313.1| serine carboxypeptidase I [Arabidopsis thaliana] 231 4e-60
gb|AAF44708.1|AF242849_1 wound-inducible carboxypeptidase [Lycop... 228 4e-59
ref|NP_189169.1| serine carboxypeptidase, putative; protein id: ... 226 2e-58
sp|P07519|CBP1_HORVU Serine carboxypeptidase I precursor (Carbox... 216 2e-55
>gb|AAM97031.1| serine carboxypeptidase 1 precursor-like protein [Arabidopsis
thaliana] gi|23197946|gb|AAN15500.1| serine
carboxypeptidase 1 precursor-like protein [Arabidopsis
thaliana]
Length = 497
Score = 234 bits (596), Expect = 8e-61
Identities = 101/144 (70%), Positives = 122/144 (84%)
Frame = -1
Query: 596 VPCTDDVVAKLWLNNEAVRKAIHTAKTSLVSQWDLCTGRIRYNHDAGSMIKYHKHLTSQG 417
VPC DD VA WLN+ +RKAIHT + S + +W+LC+G++ + HDAGSMI +H++LT G
Sbjct: 351 VPCIDDRVATAWLNDPEIRKAIHTKEESEIGRWELCSGKLSFYHDAGSMIDFHRNLTLSG 410
Query: 416 YRALIYSGDHDMCVPFTGSEAWTRSMGYKIVDEWRPWFSNDPVAGFTQGYDKNLTFMTIK 237
YRALIYSGDHDMCVPFTGSEAWT+S+GYK++DEWR W SND VAG+TQGY NLTF+TIK
Sbjct: 411 YRALIYSGDHDMCVPFTGSEAWTKSLGYKVIDEWRAWISNDQVAGYTQGYANNLTFLTIK 470
Query: 236 GAGHTVPEYKPREALEFYTHFLTG 165
GAGHTVPEYKPREAL+FY+ FL G
Sbjct: 471 GAGHTVPEYKPREALDFYSRFLEG 494
>dbj|BAB01313.1| serine carboxypeptidase I [Arabidopsis thaliana]
Length = 504
Score = 231 bits (590), Expect = 4e-60
Identities = 98/144 (68%), Positives = 119/144 (82%)
Frame = -1
Query: 596 VPCTDDVVAKLWLNNEAVRKAIHTAKTSLVSQWDLCTGRIRYNHDAGSMIKYHKHLTSQG 417
VPC DD VA WLN+ AVRKA+H + + W+LC+ + Y HD GSMI+YH++LT G
Sbjct: 358 VPCIDDTVATKWLNDPAVRKAVHAKEEKAIGNWELCSSNLEYRHDTGSMIEYHRNLTLSG 417
Query: 416 YRALIYSGDHDMCVPFTGSEAWTRSMGYKIVDEWRPWFSNDPVAGFTQGYDKNLTFMTIK 237
+RALI+SGDHDMCVP+TGSEAWT++MGYK+VDEWRPW SN+ VAGFTQGY NLTF+TIK
Sbjct: 418 FRALIFSGDHDMCVPYTGSEAWTKAMGYKVVDEWRPWMSNNQVAGFTQGYANNLTFLTIK 477
Query: 236 GAGHTVPEYKPREALEFYTHFLTG 165
GAGHTVPEYKPRE+L+FY+ FL G
Sbjct: 478 GAGHTVPEYKPRESLDFYSRFLAG 501
>gb|AAF44708.1|AF242849_1 wound-inducible carboxypeptidase [Lycopersicon esculentum]
Length = 498
Score = 228 bits (581), Expect = 4e-59
Identities = 101/144 (70%), Positives = 118/144 (81%)
Frame = -1
Query: 596 VPCTDDVVAKLWLNNEAVRKAIHTAKTSLVSQWDLCTGRIRYNHDAGSMIKYHKHLTSQG 417
VPCTDD VA LWLNN VRKAIH +++ W+LCT +I +HD+GSMI YHK+LT++G
Sbjct: 352 VPCTDDRVATLWLNNADVRKAIHAEPATVIGPWELCTDKIDLDHDSGSMIPYHKNLTARG 411
Query: 416 YRALIYSGDHDMCVPFTGSEAWTRSMGYKIVDEWRPWFSNDPVAGFTQGYDKNLTFMTIK 237
YRA+I+SGDHDMCVPFTGS WT+S+GY IVDEWRPW+ ND VAGF QGY NL FMTIK
Sbjct: 412 YRAIIFSGDHDMCVPFTGSAVWTKSLGYPIVDEWRPWYVNDQVAGFIQGYANNLIFMTIK 471
Query: 236 GAGHTVPEYKPREALEFYTHFLTG 165
GAGHTVPEYKPREAL FY+ +L G
Sbjct: 472 GAGHTVPEYKPREALAFYSRWLEG 495
>ref|NP_189169.1| serine carboxypeptidase, putative; protein id: At3g25420.1
[Arabidopsis thaliana]
Length = 505
Score = 226 bits (576), Expect = 2e-58
Identities = 99/155 (63%), Positives = 121/155 (77%), Gaps = 11/155 (7%)
Frame = -1
Query: 596 VPCTDDVVAKLWLNNEAVRKAIHTAKTSL-----------VSQWDLCTGRIRYNHDAGSM 450
VPC DD VA WLN+ AVRKA+H + S+ + W+LC+ + Y HD GSM
Sbjct: 348 VPCIDDTVATKWLNDPAVRKAVHAKEVSIQFIIFLSISISIGNWELCSSNLEYRHDTGSM 407
Query: 449 IKYHKHLTSQGYRALIYSGDHDMCVPFTGSEAWTRSMGYKIVDEWRPWFSNDPVAGFTQG 270
I+YH++LT G+RALI+SGDHDMCVP+TGSEAWT++MGYK+VDEWRPW SN+ VAGFTQG
Sbjct: 408 IEYHRNLTLSGFRALIFSGDHDMCVPYTGSEAWTKAMGYKVVDEWRPWMSNNQVAGFTQG 467
Query: 269 YDKNLTFMTIKGAGHTVPEYKPREALEFYTHFLTG 165
Y NLTF+TIKGAGHTVPEYKPRE+L+FY+ FL G
Sbjct: 468 YANNLTFLTIKGAGHTVPEYKPRESLDFYSRFLAG 502
>sp|P07519|CBP1_HORVU Serine carboxypeptidase I precursor (Carboxypeptidase C) (CP-MI)
gi|7428202|pir||CPBHS carboxypeptidase C (EC 3.4.16.5)
precursor - barley gi|1731988|emb|CAA70816.1| serine
carboxypeptidase I, CP-MI [Hordeum vulgare subsp.
vulgare]
Length = 499
Score = 216 bits (549), Expect = 2e-55
Identities = 95/147 (64%), Positives = 114/147 (76%)
Frame = -1
Query: 596 VPCTDDVVAKLWLNNEAVRKAIHTAKTSLVSQWDLCTGRIRYNHDAGSMIKYHKHLTSQG 417
VPC D VA WL+N AVR AIH S + W LCT ++ + HDAGSMI YHK+LTSQG
Sbjct: 353 VPCMSDEVATAWLDNAAVRSAIHAQSVSAIGPWLLCTDKLYFVHDAGSMIAYHKNLTSQG 412
Query: 416 YRALIYSGDHDMCVPFTGSEAWTRSMGYKIVDEWRPWFSNDPVAGFTQGYDKNLTFMTIK 237
YRA+I+SGDHDMCVPFTGSEAWT+S+GY +VD WRPW +N V+G+T+GY+ LTF TIK
Sbjct: 413 YRAIIFSGDHDMCVPFTGSEAWTKSLGYGVVDSWRPWITNGQVSGYTEGYEHGLTFATIK 472
Query: 236 GAGHTVPEYKPREALEFYTHFLTGLPL 156
GAGHTVPEYKP+EA FY+ +L G L
Sbjct: 473 GAGHTVPEYKPQEAFAFYSRWLAGSKL 499
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 530,307,465
Number of Sequences: 1393205
Number of extensions: 12347883
Number of successful extensions: 25225
Number of sequences better than 10.0: 201
Number of HSP's better than 10.0 without gapping: 24494
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25063
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)