Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
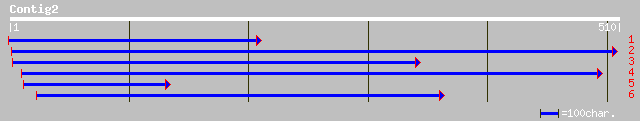
Query= KMC001038A_C02 KMC001038A_c02
(486 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB09881.1| pyrophosphate-dependent phosphofructo-1-kinase-l... 110 6e-24
ref|NP_568842.1| pyrophosphate-dependent phosphofructo-1-kinase-... 110 6e-24
ref|NP_567742.1| pyrophosphate-dependent phosphofructo-1-kinase;... 107 7e-23
pir||T06011 probable diphosphate-fructose-6-phosphate 1-phosphot... 107 7e-23
ref|NP_195010.1| putative pyrophosphate--fructose-6-phosphate 1-... 92 3e-18
>dbj|BAB09881.1| pyrophosphate-dependent phosphofructo-1-kinase-like protein
[Arabidopsis thaliana]
Length = 488
Score = 110 bits (276), Expect = 6e-24
Identities = 54/74 (72%), Positives = 61/74 (81%)
Frame = -1
Query: 486 AMPGYTGYTNGLVNGRQTYIPFYRITEKTHHVVITDRMWARLLSSTNQPSFMDPKSAHED 307
AM GYTGYT+GLVNGRQTYIPFYRITE ++VVITDRMWARLLSSTNQPSF+ PK E+
Sbjct: 403 AMAGYTGYTSGLVNGRQTYIPFYRITETQNNVVITDRMWARLLSSTNQPSFLGPKDTSEE 462
Query: 306 HKEVKPLDHPFLDD 265
KE+ + P LDD
Sbjct: 463 KKELP--ETPLLDD 474
>ref|NP_568842.1| pyrophosphate-dependent phosphofructo-1-kinase-like protein;
protein id: At5g56630.1, supported by cDNA: gi_13430589,
supported by cDNA: gi_14532861 [Arabidopsis thaliana]
gi|13430590|gb|AAK25917.1|AF360207_1 putative
pyrophosphate-dependent phosphofructo-1-kinase
[Arabidopsis thaliana] gi|14532862|gb|AAK64113.1|
putative pyrophosphate-dependent phosphofructo-1-kinase
[Arabidopsis thaliana]
Length = 485
Score = 110 bits (276), Expect = 6e-24
Identities = 54/74 (72%), Positives = 61/74 (81%)
Frame = -1
Query: 486 AMPGYTGYTNGLVNGRQTYIPFYRITEKTHHVVITDRMWARLLSSTNQPSFMDPKSAHED 307
AM GYTGYT+GLVNGRQTYIPFYRITE ++VVITDRMWARLLSSTNQPSF+ PK E+
Sbjct: 400 AMAGYTGYTSGLVNGRQTYIPFYRITETQNNVVITDRMWARLLSSTNQPSFLGPKDTSEE 459
Query: 306 HKEVKPLDHPFLDD 265
KE+ + P LDD
Sbjct: 460 KKELP--ETPLLDD 471
>ref|NP_567742.1| pyrophosphate-dependent phosphofructo-1-kinase; protein id:
At4g26270.1, supported by cDNA: gi_15146207, supported
by cDNA: gi_19699123 [Arabidopsis thaliana]
gi|15146208|gb|AAK83587.1| AT4g26270/T25K17_80
[Arabidopsis thaliana] gi|19699124|gb|AAL90928.1|
AT4g26270/T25K17_80 [Arabidopsis thaliana]
Length = 489
Score = 107 bits (267), Expect = 7e-23
Identities = 56/74 (75%), Positives = 61/74 (81%)
Frame = -1
Query: 486 AMPGYTGYTNGLVNGRQTYIPFYRITEKTHHVVITDRMWARLLSSTNQPSFMDPKSAHED 307
AM GYTGY +GLVNGRQTYIPFYRITEK +HVVITDRMWARLLSSTNQPSF+ PK D
Sbjct: 401 AMAGYTGYISGLVNGRQTYIPFYRITEKQNHVVITDRMWARLLSSTNQPSFLGPKDVF-D 459
Query: 306 HKEVKPLDHPFLDD 265
+KE KP+ LDD
Sbjct: 460 NKE-KPMS-ALLDD 471
>pir||T06011 probable diphosphate-fructose-6-phosphate 1-phosphotransferase (EC
2.7.1.90) T25K17.80 - Arabidopsis thaliana
gi|4539423|emb|CAB38956.1| pyrophosphate-dependent
phosphofructo-1-kinase [Arabidopsis thaliana]
gi|7269478|emb|CAB79482.1| pyrophosphate-dependent
phosphofructo-1-kinase [Arabidopsis thaliana]
Length = 500
Score = 107 bits (267), Expect = 7e-23
Identities = 56/74 (75%), Positives = 61/74 (81%)
Frame = -1
Query: 486 AMPGYTGYTNGLVNGRQTYIPFYRITEKTHHVVITDRMWARLLSSTNQPSFMDPKSAHED 307
AM GYTGY +GLVNGRQTYIPFYRITEK +HVVITDRMWARLLSSTNQPSF+ PK D
Sbjct: 412 AMAGYTGYISGLVNGRQTYIPFYRITEKQNHVVITDRMWARLLSSTNQPSFLGPKDVF-D 470
Query: 306 HKEVKPLDHPFLDD 265
+KE KP+ LDD
Sbjct: 471 NKE-KPMS-ALLDD 482
>ref|NP_195010.1| putative pyrophosphate--fructose-6-phosphate 1-phosphotransferase;
protein id: At4g32840.1, supported by cDNA: 40445.
[Arabidopsis thaliana] gi|7434215|pir||T10691 probable
diphosphate-fructose-6-phosphate 1-phosphotransferase
(EC 2.7.1.90) T16I18.50 - Arabidopsis thaliana
gi|7270231|emb|CAB80001.1| putative
pyrophosphate--fructose-6-phosphate 1-phosphotransferase
[Arabidopsis thaliana] gi|29029114|gb|AAO64936.1|
At4g32840 [Arabidopsis thaliana]
Length = 462
Score = 92.0 bits (227), Expect = 3e-18
Identities = 42/54 (77%), Positives = 48/54 (88%)
Frame = -1
Query: 486 AMPGYTGYTNGLVNGRQTYIPFYRITEKTHHVVITDRMWARLLSSTNQPSFMDP 325
AM GYTG+ +GLVNGR TYIPF RITE+ + VVITDRMWAR+LSSTNQPSFM+P
Sbjct: 400 AMAGYTGFVSGLVNGRHTYIPFNRITERQNKVVITDRMWARMLSSTNQPSFMNP 453
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 456,796,514
Number of Sequences: 1393205
Number of extensions: 9986136
Number of successful extensions: 22276
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 21503
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22274
length of database: 448,689,247
effective HSP length: 113
effective length of database: 291,257,082
effective search space used: 13980339936
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)