Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001019A_C01 KMC001019A_c01
(559 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
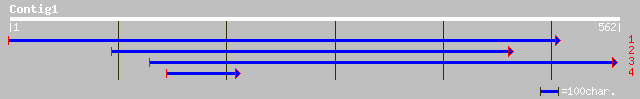
ref|NP_178143.1| hypothetical protein; protein id: At1g80270.1, ... 134 1e-30
ref|NP_173001.1| hypothetical protein; protein id: At1g15480.1 [... 128 5e-29
ref|NP_188178.2| DNA-binding protein, putative; protein id: At3g... 123 2e-27
dbj|BAA87841.1| unnamed protein product [Oryza sativa (japonica ... 121 7e-27
gb|AAF32491.1|AF091837_1 DNA-binding protein [Triticum aestivum] 119 2e-26
>ref|NP_178143.1| hypothetical protein; protein id: At1g80270.1, supported by cDNA:
gi_17064897, supported by cDNA: gi_20259917 [Arabidopsis
thaliana] gi|25406647|pir||B96834 hypothetical protein
F5I6.2 [imported] - Arabidopsis thaliana
gi|12324975|gb|AAG52431.1|AC018848_2 hypothetical
protein; 8785-10851 [Arabidopsis thaliana]
gi|17064898|gb|AAL32603.1| Unknown protein [Arabidopsis
thaliana] gi|20259918|gb|AAM13306.1| unknown protein
[Arabidopsis thaliana]
Length = 596
Score = 134 bits (336), Expect = 1e-30
Identities = 62/109 (56%), Positives = 87/109 (78%)
Frame = -2
Query: 558 SIFQKATQPSHAHQVNPLFFSYIAILEQYSKRGDIHNSEKIFYRMKQAGYTSRIRQYQVL 379
S+ KA++ SH + S++ I+++YSKRGD+HN+EKIF +M++AGYTSR+RQ+Q L
Sbjct: 491 SLLDKASKQSHTKL---MMNSFMYIMDEYSKRGDVHNTEKIFLKMREAGYTSRLRQFQAL 547
Query: 378 LQAYIKAKLPAYGIRDRLKGDNIYPNRNLATLLAQVDGFRKTPVSDLLD 232
+QAYI AK PAYG+RDRLK DNI+PN+++A LAQ D F+KT +SD+LD
Sbjct: 548 MQAYINAKSPAYGMRDRLKADNIFPNKSMAAQLAQGDPFKKTAISDILD 596
>ref|NP_173001.1| hypothetical protein; protein id: At1g15480.1 [Arabidopsis
thaliana] gi|5103846|gb|AAD39676.1|AC007591_41 F9L1.43
[Arabidopsis thaliana]
Length = 623
Score = 128 bits (321), Expect = 5e-29
Identities = 60/109 (55%), Positives = 81/109 (74%)
Frame = -2
Query: 558 SIFQKATQPSHAHQVNPLFFSYIAILEQYSKRGDIHNSEKIFYRMKQAGYTSRIRQYQVL 379
S KA Q + Q+ PL S++ ++ +Y +RGD+HN+EKIF RMKQAGY SR YQ L
Sbjct: 518 SSLSKAIQ---SKQIKPLMSSFMYLMHEYVRRGDVHNTEKIFQRMKQAGYQSRFWAYQTL 574
Query: 378 LQAYIKAKLPAYGIRDRLKGDNIYPNRNLATLLAQVDGFRKTPVSDLLD 232
+QAY+ AK PAYG+++R+K DNI+PN+ LA LA+ D F+KTP+SDLLD
Sbjct: 575 IQAYVNAKAPAYGMKERMKADNIFPNKRLAAQLAKADPFKKTPLSDLLD 623
>ref|NP_188178.2| DNA-binding protein, putative; protein id: At3g15590.1, supported
by cDNA: gi_20856657 [Arabidopsis thaliana]
gi|11994226|dbj|BAB01348.1| DNA-binding protein
[Arabidopsis thaliana] gi|20856658|gb|AAM26678.1|
AT3g15590/MQD17_5 [Arabidopsis thaliana]
gi|27363266|gb|AAO11552.1| At3g15590/MQD17_5
[Arabidopsis thaliana]
Length = 610
Score = 123 bits (308), Expect = 2e-27
Identities = 56/108 (51%), Positives = 85/108 (77%)
Frame = -2
Query: 555 IFQKATQPSHAHQVNPLFFSYIAILEQYSKRGDIHNSEKIFYRMKQAGYTSRIRQYQVLL 376
I +AT+ + ++ P+F +Y+AILE+Y+KRGD+HN+EK+F +MK+A Y +++ QY+ +L
Sbjct: 505 ILNRATKDN---KMRPMFTTYMAILEEYAKRGDVHNTEKVFMKMKRASYAAQLMQYETVL 561
Query: 375 QAYIKAKLPAYGIRDRLKGDNIYPNRNLATLLAQVDGFRKTPVSDLLD 232
AYI AK PAYG+ +R+K DN++PN++LA LAQV+ F+K PVS LLD
Sbjct: 562 LAYINAKTPAYGMIERMKADNVFPNKSLAAKLAQVNPFKKCPVSVLLD 609
>dbj|BAA87841.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|11138066|dbj|BAB17739.1| putative DNA-binding protein
[Oryza sativa (japonica cultivar-group)]
gi|13873009|dbj|BAB44113.1| putative DNA-binding protein
[Oryza sativa (japonica cultivar-group)]
Length = 591
Score = 121 bits (303), Expect = 7e-27
Identities = 53/101 (52%), Positives = 81/101 (79%), Gaps = 1/101 (0%)
Frame = -2
Query: 531 SHAHQVNPLFFSYIAILEQYSKRGDIHNSEKIFYRMKQAGYTSRIRQYQVLLQAYIKAKL 352
S+ +++ PL+ +Y+ +L+ YSK+GD+HN+EK+F +++Q GYT RIRQYQ+LL+AY+ AK
Sbjct: 491 SYKNKIKPLYTTYLMLLDSYSKKGDVHNAEKLFSKVRQMGYTGRIRQYQLLLEAYLNAKT 550
Query: 351 PAYGIRDRLKGDNIYPNRNLATLLAQVDGF-RKTPVSDLLD 232
P YG ++R+K D+I+PNR +A+LLA D F RK +S+LLD
Sbjct: 551 PPYGFKERMKADDIFPNRAVASLLAATDPFNRKNAMSELLD 591
Score = 33.5 bits (75), Expect = 1.8
Identities = 16/56 (28%), Positives = 27/56 (47%)
Frame = -2
Query: 519 QVNPLFFSYIAILEQYSKRGDIHNSEKIFYRMKQAGYTSRIRQYQVLLQAYIKAKL 352
+VN ++ +E + K GD+ +E+IF M + T Y +L+ Y KL
Sbjct: 389 EVNARLDECMSAIEAFGKLGDVEKAEEIFENMFKTWKTLSFEYYNAMLKVYANKKL 444
>gb|AAF32491.1|AF091837_1 DNA-binding protein [Triticum aestivum]
Length = 612
Score = 119 bits (299), Expect = 2e-26
Identities = 52/100 (52%), Positives = 76/100 (76%)
Frame = -2
Query: 531 SHAHQVNPLFFSYIAILEQYSKRGDIHNSEKIFYRMKQAGYTSRIRQYQVLLQAYIKAKL 352
S ++++ P + SY+ +L+ YSK+GDIHNSEK+F +++Q GY R+RQYQ+LL AY+ AK
Sbjct: 513 SKSNKMKPQYSSYLMLLDTYSKKGDIHNSEKVFDQLRQMGYNGRVRQYQLLLNAYVHAKT 572
Query: 351 PAYGIRDRLKGDNIYPNRNLATLLAQVDGFRKTPVSDLLD 232
P YG R+R+K DNI+PN +A+LLA D F K +SD+L+
Sbjct: 573 PVYGFRERMKADNIFPNSVIASLLAATDPFNKKTLSDMLE 612
Score = 31.2 bits (69), Expect = 9.0
Identities = 14/56 (25%), Positives = 26/56 (46%)
Frame = -2
Query: 519 QVNPLFFSYIAILEQYSKRGDIHNSEKIFYRMKQAGYTSRIRQYQVLLQAYIKAKL 352
+ NP ++ +E + + GD+ +EK+F M T + Y L++ Y L
Sbjct: 411 EANPRLDECLSAIESFGRLGDVERAEKVFEDMFATWKTLSSKFYNALMKVYADQNL 466
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 470,077,330
Number of Sequences: 1393205
Number of extensions: 9657437
Number of successful extensions: 21701
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 20906
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21700
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)