Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001015A_C02 KMC001015A_c02
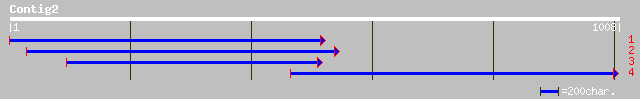
(1007 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_133474.2| similar to FLJ00034 protein [Homo sapiens] [Mus... 43 0.007
sp|P98089|MUCL_RAT Intestinal mucin-like protein (MLP) 42 0.020
pir||A42112 mucin-like peptide MLP 2677 - rat 42 0.020
ref|NP_062257.1| CTD-binding SR-like [Rattus norvegicus] gi|7513... 42 0.020
ref|NP_190627.1| proline-rich protein; protein id: At3g50580.1 [... 41 0.026
>ref|XP_133474.2| similar to FLJ00034 protein [Homo sapiens] [Mus musculus]
Length = 350
Score = 43.1 bits (100), Expect = 0.007
Identities = 30/67 (44%), Positives = 38/67 (55%), Gaps = 8/67 (11%)
Frame = +1
Query: 43 SYTASP-CSKDCPL---DRFPAPPPSGTPSSSASSATRSPSTPSFSTPT*EPPLPPPT-- 204
S+ + P CS+ L D PAPPP+ PSS++SS + SPS+ S S P PP PPP
Sbjct: 165 SHASRPACSRHLTLGTGDGGPAPPPA--PSSASSSPSPSPSSSSPSPPPPPPPPPPPALP 222
Query: 205 --HFDSY 219
FD Y
Sbjct: 223 APRFDIY 229
>sp|P98089|MUCL_RAT Intestinal mucin-like protein (MLP)
Length = 837
Score = 41.6 bits (96), Expect = 0.020
Identities = 24/64 (37%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Frame = +1
Query: 61 CSKDCPLDRF-------PAPPPSGTPSSSASSATRSPSTPSFSTPT*EPPLPPPTHFDSY 219
CS DC L F P+ PP TP++ SS T +PSTPS ++ P P T S
Sbjct: 3 CSVDCQLQVFNWSCPSTPSTPPPSTPTTPTSSQTTTPSTPSTTSSKSTPSTPQSTSSKST 62
Query: 220 REPP 231
P
Sbjct: 63 PSTP 66
>pir||A42112 mucin-like peptide MLP 2677 - rat
Length = 837
Score = 41.6 bits (96), Expect = 0.020
Identities = 24/64 (37%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Frame = +1
Query: 61 CSKDCPLDRF-------PAPPPSGTPSSSASSATRSPSTPSFSTPT*EPPLPPPTHFDSY 219
CS DC L F P+ PP TP++ SS T +PSTPS ++ P P T S
Sbjct: 3 CSVDCQLQVFNWSCPSTPSTPPPSTPTTPTSSQTTTPSTPSTTSSKSTPSTPQSTSSKST 62
Query: 220 REPP 231
P
Sbjct: 63 PSTP 66
>ref|NP_062257.1| CTD-binding SR-like [Rattus norvegicus] gi|7513923|pir||T31421
C-terminal domain-binding protein rA1 - rat
gi|1438532|gb|AAC52657.1| rA1
Length = 1173
Score = 41.6 bits (96), Expect = 0.020
Identities = 25/50 (50%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Frame = +1
Query: 82 DRFPAPPPSGTPSSSASSATRSPSTPSFSTPT*EPPLPPPT----HFDSY 219
D PAPPP+ PSS +SS + SPS+ S S P PP PPP FD Y
Sbjct: 98 DGGPAPPPA--PSSGSSSPSPSPSSSSPSPPPPPPPPPPPALPAPRFDIY 145
>ref|NP_190627.1| proline-rich protein; protein id: At3g50580.1 [Arabidopsis
thaliana] gi|11358710|pir||T46089 proline-rich protein -
Arabidopsis thaliana gi|6561998|emb|CAB62487.1|
proline-rich protein [Arabidopsis thaliana]
Length = 265
Score = 41.2 bits (95), Expect = 0.026
Identities = 23/49 (46%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Frame = +1
Query: 91 PAPPPSGTPSSSASSATRSPSTPSFSTPT*E--PPLPPPTHFDSYREPP 231
P+PPP TPS + +SPSTPS PT + PP PP H S PP
Sbjct: 126 PSPPP--TPSLPPPAPKKSPSTPSLPPPTPKKSPPPPPSHHSSSPSNPP 172
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 923,141,611
Number of Sequences: 1393205
Number of extensions: 23124782
Number of successful extensions: 138537
Number of sequences better than 10.0: 515
Number of HSP's better than 10.0 without gapping: 86708
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 125279
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 58221615497
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)