Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001011A_C01 KMC001011A_c01
(690 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
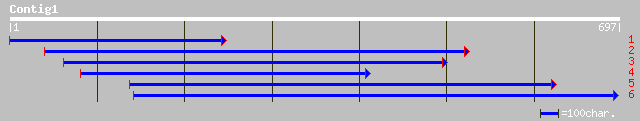
ref|NP_174096.1| hypothetical protein; protein id: At1g27750.1 [... 146 2e-34
gb|AAM13844.1| unknown protein [Arabidopsis thaliana] 146 2e-34
gb|AAF24949.1|AC012375_12 T22C5.20 [Arabidopsis thaliana] 118 9e-26
dbj|BAB63599.1| P0403C05.21 [Oryza sativa (japonica cultivar-gro... 48 1e-04
gb|AAM00273.1|AF363506_1 lipid transfer protein 2 [Euphorbia lag... 33 3.7
>ref|NP_174096.1| hypothetical protein; protein id: At1g27750.1 [Arabidopsis thaliana]
Length = 1975
Score = 146 bits (369), Expect = 2e-34
Identities = 66/102 (64%), Positives = 82/102 (79%)
Frame = -3
Query: 670 PEPAEWPTQLDMTIRTDFRHVKSTLAATPSHRREVCHLIPSSTSDHERFQDFVSYLKQKD 491
PEP +WP +LDMT RTD ++VK+T A T H+REVC LIP++ SD + QDF++YLKQ+D
Sbjct: 961 PEPVQWPVKLDMTKRTDMKNVKATFANTQPHKREVCQLIPAAFSDRKGLQDFITYLKQRD 1020
Query: 490 CAGVINIPASKSIWARLLFILPHSLETCSLLSIAPDPSDCLI 365
CAGVI IPAS +WAR LFILPHS ETCSLLS++P S+CLI
Sbjct: 1021 CAGVIKIPASSPMWARHLFILPHSQETCSLLSVSPSSSECLI 1062
>gb|AAM13844.1| unknown protein [Arabidopsis thaliana]
Length = 1075
Score = 146 bits (369), Expect = 2e-34
Identities = 66/102 (64%), Positives = 82/102 (79%)
Frame = -3
Query: 670 PEPAEWPTQLDMTIRTDFRHVKSTLAATPSHRREVCHLIPSSTSDHERFQDFVSYLKQKD 491
PEP +WP +LDMT RTD ++VK+T A T H+REVC LIP++ SD + QDF++YLKQ+D
Sbjct: 961 PEPVQWPVKLDMTKRTDMKNVKATFANTQPHKREVCQLIPAAFSDRKGLQDFITYLKQRD 1020
Query: 490 CAGVINIPASKSIWARLLFILPHSLETCSLLSIAPDPSDCLI 365
CAGVI IPAS +WAR LFILPHS ETCSLLS++P S+CLI
Sbjct: 1021 CAGVIKIPASSPMWARHLFILPHSQETCSLLSVSPSSSECLI 1062
>gb|AAF24949.1|AC012375_12 T22C5.20 [Arabidopsis thaliana]
Length = 1840
Score = 118 bits (295), Expect = 9e-26
Identities = 57/102 (55%), Positives = 70/102 (67%)
Frame = -3
Query: 670 PEPAEWPTQLDMTIRTDFRHVKSTLAATPSHRREVCHLIPSSTSDHERFQDFVSYLKQKD 491
PEP +WP +LDMT RTD ++VK+T A T H+ QDF++YLKQ+D
Sbjct: 944 PEPVQWPVKLDMTKRTDMKNVKATFANTQPHK----------------LQDFITYLKQRD 987
Query: 490 CAGVINIPASKSIWARLLFILPHSLETCSLLSIAPDPSDCLI 365
CAGVI IPAS +WAR LFILPHS ETCSLLS++P S+CLI
Sbjct: 988 CAGVIKIPASSPMWARHLFILPHSQETCSLLSVSPSSSECLI 1029
>dbj|BAB63599.1| P0403C05.21 [Oryza sativa (japonica cultivar-group)]
Length = 798
Score = 47.8 bits (112), Expect = 1e-04
Identities = 18/35 (51%), Positives = 27/35 (76%)
Frame = -3
Query: 688 RYSNTIPEPAEWPTQLDMTIRTDFRHVKSTLAATP 584
RY + + EP WP++LD+T RTD++HVK+T + TP
Sbjct: 758 RYEHAVSEPTGWPSRLDVTKRTDYQHVKTTFSNTP 792
>gb|AAM00273.1|AF363506_1 lipid transfer protein 2 [Euphorbia lagascae]
Length = 116
Score = 33.1 bits (74), Expect = 3.7
Identities = 18/48 (37%), Positives = 26/48 (53%)
Frame = -3
Query: 598 LAATPSHRREVCHLIPSSTSDHERFQDFVSYLKQKDCAGVINIPASKS 455
+A T + RR C + ++ S + + S L KDC VINIP SK+
Sbjct: 63 IAQTTADRRAACECVKAAASHYTINEKAASSLP-KDCGAVINIPISKT 109
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 586,994,350
Number of Sequences: 1393205
Number of extensions: 12557815
Number of successful extensions: 25448
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 24744
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25436
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31118763720
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)