Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001004A_C01 KMC001004A_c01
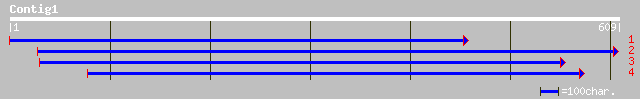
(606 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_609026.1| CG9523-PA [Drosophila melanogaster] gi|20152055... 33 2.9
gb|AAA82711.1| hypothetical OrfZ 32 6.4
gb|AAA64917.1| ORFZ 32 6.4
ref|NP_230493.1| conserved hypothetical protein [Vibrio cholerae... 32 6.4
gb|AAK28043.1|AF304131_1 transmembrane receptor Roundabout3 [Dan... 32 8.4
>ref|NP_609026.1| CG9523-PA [Drosophila melanogaster] gi|20152055|gb|AAM11387.1|
LD47713p [Drosophila melanogaster]
gi|22945765|gb|AAF52381.2| CG9523-PA [Drosophila
melanogaster]
Length = 492
Score = 33.1 bits (74), Expect = 2.9
Identities = 18/54 (33%), Positives = 32/54 (58%), Gaps = 1/54 (1%)
Frame = +2
Query: 314 PEQQKLEQVYMLRIVNSGQVHIFPSSFVSQATPSTVPL-LNETVKLPRKIPLLL 472
P+QQ+ + + L++ N G + F F++ T T+ L L T LP++IP+L+
Sbjct: 404 PKQQRSKYYHFLKLANEGDIRPF-VRFIADCTEKTLDLYLWATSDLPQQIPMLI 456
>gb|AAA82711.1| hypothetical OrfZ
Length = 191
Score = 32.0 bits (71), Expect = 6.4
Identities = 19/52 (36%), Positives = 26/52 (49%)
Frame = +2
Query: 296 RHFLDFPEQQKLEQVYMLRIVNSGQVHIFPSSFVSQATPSTVPLLNETVKLP 451
RH LD P+ Q LE + +I N QV PS+ V +PS P + + P
Sbjct: 94 RHNLDLPKDQFLEAITE-KIANDNQVDKQPSTHVPNISPSFTPGTSSNLNSP 144
>gb|AAA64917.1| ORFZ
Length = 194
Score = 32.0 bits (71), Expect = 6.4
Identities = 19/52 (36%), Positives = 26/52 (49%)
Frame = +2
Query: 296 RHFLDFPEQQKLEQVYMLRIVNSGQVHIFPSSFVSQATPSTVPLLNETVKLP 451
RH LD P+ Q LE + +I N QV PS+ V +PS P + + P
Sbjct: 97 RHNLDLPKDQFLEAITE-KIANDNQVDKQPSTHVPNISPSFTPGTSSNLNSP 147
>ref|NP_230493.1| conserved hypothetical protein [Vibrio cholerae]
gi|20177824|sp|Q9KTQ4|ACFD_VIBCH Accessory colonization
factor AcfD precursor gi|11354556|pir||B82274 conserved
hypothetical protein VC0845 [imported] - Vibrio cholerae
(strain N16961 serogroup O1) gi|9655297|gb|AAF94008.1|
conserved hypothetical protein [Vibrio cholerae]
gi|13377567|gb|AAK20802.1|AF325734_28 accessory
colonization factor AcfD [Vibrio cholerae]
Length = 1520
Score = 32.0 bits (71), Expect = 6.4
Identities = 19/52 (36%), Positives = 26/52 (49%)
Frame = +2
Query: 296 RHFLDFPEQQKLEQVYMLRIVNSGQVHIFPSSFVSQATPSTVPLLNETVKLP 451
RH LD P+ Q LE + +I N QV PS+ V +PS P + + P
Sbjct: 171 RHNLDLPKDQFLEAITE-KIANDNQVDKQPSTHVPNISPSFTPGTSSNLNSP 221
>gb|AAK28043.1|AF304131_1 transmembrane receptor Roundabout3 [Danio rerio]
Length = 1419
Score = 31.6 bits (70), Expect = 8.4
Identities = 17/45 (37%), Positives = 25/45 (54%), Gaps = 2/45 (4%)
Frame = +2
Query: 287 RTQRHFLDFPEQQKLEQVYMLRIVNSGQVHI--FPSSFVSQATPS 415
R HF D P +LEQ +M R + G VH+ PS ++Q+ P+
Sbjct: 1194 RDDGHFHDIPRLHQLEQPHMTRRLPCGPVHVPRAPSPPLTQSNPN 1238
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 456,718,402
Number of Sequences: 1393205
Number of extensions: 8908061
Number of successful extensions: 21319
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 20604
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21302
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)