Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001003A_C01 KMC001003A_c01
(761 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
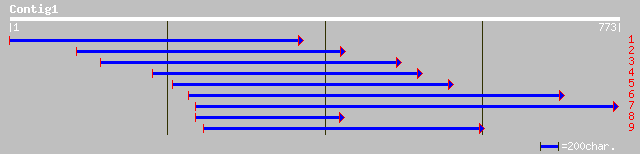
gb|AAO66528.1| putative CEO protein [Oryza sativa (japonica cult... 96 7e-19
gb|AAL24144.1| unknown protein [Arabidopsis thaliana] gi|2329687... 92 8e-18
ref|NP_564391.1| expressed protein; protein id: At1g32230.1, sup... 92 8e-18
pir||H86446 unknown protein [imported] - Arabidopsis thaliana gi... 92 8e-18
gb|AAK54509.1|AF317898_1 ATP8 [Arabidopsis thaliana] 92 8e-18
>gb|AAO66528.1| putative CEO protein [Oryza sativa (japonica cultivar-group)]
Length = 584
Score = 95.5 bits (236), Expect = 7e-19
Identities = 45/79 (56%), Positives = 58/79 (72%)
Frame = -3
Query: 741 ASSHKAPRSPWMPFPMLFAAITEKVPSHDMNLTHMHYQPFRSKKITRDDFVKKLRLIVGD 562
A S ++P SPWMPF MLFAAI+ KVP DM+L H +Y+ F+ +KI+R D VK+LR IVGD
Sbjct: 490 APSPRSPSSPWMPFSMLFAAISAKVPRSDMDLVHKYYEEFKRRKISRPDLVKQLRQIVGD 549
Query: 561 TLLRATITSLQCKIPPQTA 505
LL +T+ LQ K+PP A
Sbjct: 550 KLLVSTVVRLQQKLPPMAA 568
>gb|AAL24144.1| unknown protein [Arabidopsis thaliana] gi|23296877|gb|AAN13193.1|
unknown protein [Arabidopsis thaliana]
Length = 588
Score = 92.0 bits (227), Expect = 8e-18
Identities = 45/73 (61%), Positives = 56/73 (76%)
Frame = -3
Query: 747 TVASSHKAPRSPWMPFPMLFAAITEKVPSHDMNLTHMHYQPFRSKKITRDDFVKKLRLIV 568
+V SS P+SPWMPFP LFAAI+ KV +DM L + YQ R KK+TR +FV+KLR+IV
Sbjct: 494 SVGSSTTRPKSPWMPFPTLFAAISHKVAENDMLLINADYQQLRDKKMTRAEFVRKLRVIV 553
Query: 567 GDTLLRATITSLQ 529
GD LLR+TIT+LQ
Sbjct: 554 GDDLLRSTITTLQ 566
>ref|NP_564391.1| expressed protein; protein id: At1g32230.1, supported by cDNA:
gi_11044956, supported by cDNA: gi_19715642 [Arabidopsis
thaliana] gi|19715643|gb|AAL91641.1| At1g32230/F3C3_1
[Arabidopsis thaliana]
Length = 589
Score = 92.0 bits (227), Expect = 8e-18
Identities = 45/73 (61%), Positives = 56/73 (76%)
Frame = -3
Query: 747 TVASSHKAPRSPWMPFPMLFAAITEKVPSHDMNLTHMHYQPFRSKKITRDDFVKKLRLIV 568
+V SS P+SPWMPFP LFAAI+ KV +DM L + YQ R KK+TR +FV+KLR+IV
Sbjct: 495 SVGSSTTRPKSPWMPFPTLFAAISHKVAENDMLLINADYQQLRDKKMTRAEFVRKLRVIV 554
Query: 567 GDTLLRATITSLQ 529
GD LLR+TIT+LQ
Sbjct: 555 GDDLLRSTITTLQ 567
>pir||H86446 unknown protein [imported] - Arabidopsis thaliana
gi|10801372|gb|AAG23444.1|AC084165_10 unknown protein
[Arabidopsis thaliana]
Length = 596
Score = 92.0 bits (227), Expect = 8e-18
Identities = 45/73 (61%), Positives = 56/73 (76%)
Frame = -3
Query: 747 TVASSHKAPRSPWMPFPMLFAAITEKVPSHDMNLTHMHYQPFRSKKITRDDFVKKLRLIV 568
+V SS P+SPWMPFP LFAAI+ KV +DM L + YQ R KK+TR +FV+KLR+IV
Sbjct: 502 SVGSSTTRPKSPWMPFPTLFAAISHKVAENDMLLINADYQQLRDKKMTRAEFVRKLRVIV 561
Query: 567 GDTLLRATITSLQ 529
GD LLR+TIT+LQ
Sbjct: 562 GDDLLRSTITTLQ 574
>gb|AAK54509.1|AF317898_1 ATP8 [Arabidopsis thaliana]
Length = 571
Score = 92.0 bits (227), Expect = 8e-18
Identities = 45/73 (61%), Positives = 56/73 (76%)
Frame = -3
Query: 747 TVASSHKAPRSPWMPFPMLFAAITEKVPSHDMNLTHMHYQPFRSKKITRDDFVKKLRLIV 568
+V SS P+SPWMPFP LFAAI+ KV +DM L + YQ R KK+TR +FV+KLR+IV
Sbjct: 478 SVGSSTTRPKSPWMPFPTLFAAISHKVAENDMLLINADYQQLRDKKMTRAEFVRKLRVIV 537
Query: 567 GDTLLRATITSLQ 529
GD LLR+TIT+LQ
Sbjct: 538 GDDLLRSTITTLQ 550
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 629,364,757
Number of Sequences: 1393205
Number of extensions: 13605966
Number of successful extensions: 33149
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 31742
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33120
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 36974710344
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)