Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC001001A_C01 KMC001001A_c01
(600 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
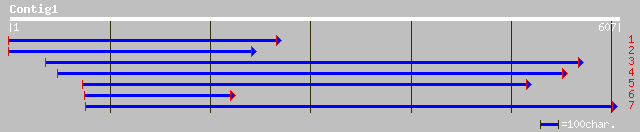
gb|AAM91579.1| unknown protein [Arabidopsis thaliana] 122 4e-27
ref|NP_175736.1| hypothetical protein; protein id: At1g53290.1 [... 122 4e-27
ref|NP_188114.1| galactosyltransferase, putative; protein id: At... 118 5e-26
emb|CAD44839.1| beta 1,3-glycosyltransferase-like protein III [O... 107 1e-22
ref|NP_180179.1| unknown protein; protein id: At2g26100.1 [Arabi... 100 2e-20
>gb|AAM91579.1| unknown protein [Arabidopsis thaliana]
Length = 345
Score = 122 bits (306), Expect = 4e-27
Identities = 52/66 (78%), Positives = 62/66 (93%)
Frame = -1
Query: 600 IGAWMLAMNVNHENNHELCAPDCTATSVAVWDIPKCSGLCNPEKKMLELHQKESCSQSPT 421
IGAWMLAMNVNHEN+H LC P+C+ +SVAVWDIPKCSGLCNPEK+MLELH++ESCS+SPT
Sbjct: 280 IGAWMLAMNVNHENHHILCEPECSPSSVAVWDIPKCSGLCNPEKRMLELHKQESCSKSPT 339
Query: 420 LEADDD 403
L +DD+
Sbjct: 340 LPSDDE 345
>ref|NP_175736.1| hypothetical protein; protein id: At1g53290.1 [Arabidopsis
thaliana] gi|25405979|pir||C96573 protein F12M16.19
[imported] - Arabidopsis thaliana
gi|7769857|gb|AAF69535.1|AC008007_10 F12M16.19
[Arabidopsis thaliana]
Length = 353
Score = 122 bits (306), Expect = 4e-27
Identities = 52/66 (78%), Positives = 62/66 (93%)
Frame = -1
Query: 600 IGAWMLAMNVNHENNHELCAPDCTATSVAVWDIPKCSGLCNPEKKMLELHQKESCSQSPT 421
IGAWMLAMNVNHEN+H LC P+C+ +SVAVWDIPKCSGLCNPEK+MLELH++ESCS+SPT
Sbjct: 288 IGAWMLAMNVNHENHHILCEPECSPSSVAVWDIPKCSGLCNPEKRMLELHKQESCSKSPT 347
Query: 420 LEADDD 403
L +DD+
Sbjct: 348 LPSDDE 353
>ref|NP_188114.1| galactosyltransferase, putative; protein id: At3g14960.1, supported
by cDNA: gi_17979336 [Arabidopsis thaliana]
gi|8777479|dbj|BAA97059.1| contains similarity to Avr9
elicitor response protein~gene_id:K15M2.10 [Arabidopsis
thaliana] gi|17979337|gb|AAL49894.1| putative
galactosyltransferase [Arabidopsis thaliana]
gi|22136686|gb|AAM91662.1| putative
galactosyltransferase [Arabidopsis thaliana]
Length = 343
Score = 118 bits (296), Expect = 5e-26
Identities = 51/66 (77%), Positives = 58/66 (87%)
Frame = -1
Query: 600 IGAWMLAMNVNHENNHELCAPDCTATSVAVWDIPKCSGLCNPEKKMLELHQKESCSQSPT 421
IGAWMLAMNVNHEN H LC P+C+ S+AVWDIPKCSGLCNPEK+MLELH ESCS+SPT
Sbjct: 278 IGAWMLAMNVNHENLHTLCEPECSPYSIAVWDIPKCSGLCNPEKRMLELHMLESCSKSPT 337
Query: 420 LEADDD 403
L +DD+
Sbjct: 338 LPSDDE 343
>emb|CAD44839.1| beta 1,3-glycosyltransferase-like protein III [Oryza sativa]
Length = 207
Score = 107 bits (267), Expect = 1e-22
Identities = 43/66 (65%), Positives = 56/66 (84%)
Frame = -1
Query: 600 IGAWMLAMNVNHENNHELCAPDCTATSVAVWDIPKCSGLCNPEKKMLELHQKESCSQSPT 421
IG+WMLAMNVNHEN H LC+P+CT +S+AVWDIPKCSGLC+PE KMLELH+++ C+ P+
Sbjct: 140 IGSWMLAMNVNHENTHALCSPECTESSIAVWDIPKCSGLCHPEVKMLELHRRKECTGGPS 199
Query: 420 LEADDD 403
++ D
Sbjct: 200 AVSESD 205
>ref|NP_180179.1| unknown protein; protein id: At2g26100.1 [Arabidopsis thaliana]
gi|7487205|pir||T02614 hypothetical protein At2g26100
[imported] - Arabidopsis thaliana
gi|3413704|gb|AAC31227.1| unknown protein [Arabidopsis
thaliana]
Length = 333
Score = 99.8 bits (247), Expect = 2e-20
Identities = 41/65 (63%), Positives = 53/65 (81%)
Frame = -1
Query: 600 IGAWMLAMNVNHENNHELCAPDCTATSVAVWDIPKCSGLCNPEKKMLELHQKESCSQSPT 421
IG+WMLAM+V+HE+N LC P C+ S+AVWDIPKCSGLC+PE ++ ELH+ + CS+SPT
Sbjct: 266 IGSWMLAMDVHHEDNRALCDPHCSPKSIAVWDIPKCSGLCDPESRLKELHKTDMCSKSPT 325
Query: 420 LEADD 406
L DD
Sbjct: 326 LPPDD 330
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 523,690,509
Number of Sequences: 1393205
Number of extensions: 11513410
Number of successful extensions: 26776
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 26064
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26763
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23426109484
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)