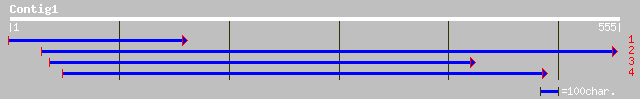
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000997A_C01 KMC000997A_c01
(551 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO63778.1| unknown [Populus tremuloides] 122 2e-27
gb|AAH06040.1| Unknown (protein for MGC:7642) [Mus musculus] 102 3e-21
pir||F86145 F22L4.5 protein - Arabidopsis thaliana gi|8920587|gb... 96 4e-19
ref|NP_171656.1| expressed protein; protein id: At1g01490.1, sup... 96 4e-19
gb|AAM88621.1| hypothetical protein [Oryza sativa (japonica cult... 78 8e-14
>gb|AAO63778.1| unknown [Populus tremuloides]
Length = 132
Score = 122 bits (307), Expect = 2e-27
Identities = 67/130 (51%), Positives = 86/130 (65%), Gaps = 12/130 (9%)
Frame = -2
Query: 550 ELHDDKIKQKAMKSVSXISGXESVSVDMGSKKLTLTGDIDPVHVVSKLRKWCHTEIVSVG 371
+LHD+K K KAMK VS +SG +S+S+DM KKLT+ GD+DPVH+VSKLRK C+TEI++VG
Sbjct: 9 DLHDEKAKTKAMKKVSSLSGVDSISMDMKDKKLTVIGDVDPVHIVSKLRKLCNTEIITVG 68
Query: 370 PAKEEKKKDESSKPDQPK------------LPEPVKFYEAYPLYYQTRPSQYTQYHYVQS 227
PAKE +KK E K ++PK + E VK Y+AY P T YH V+S
Sbjct: 69 PAKEPEKKKEEPKKEEPKKQEDPKKKEQDAVAELVKAYKAY------NPPMTTYYH-VRS 121
Query: 226 VEEDNVGCVI 197
VE+D CVI
Sbjct: 122 VEDDPNACVI 131
>gb|AAH06040.1| Unknown (protein for MGC:7642) [Mus musculus]
Length = 167
Score = 102 bits (254), Expect = 3e-21
Identities = 65/162 (40%), Positives = 86/162 (52%), Gaps = 43/162 (26%)
Frame = -2
Query: 550 ELHDDKIKQKAMKSVSXISGXESVSVDMGSKKLTLTGDIDPVHVVSKLRK-WCHTEIVSV 374
+LHDDK KQKA+K+VS + G +S+++DM +KLT+ G +DPV +VSKLRK W T+IV V
Sbjct: 9 DLHDDKAKQKALKTVSTLPGIDSIAMDMKERKLTVIGSVDPVSIVSKLRKYWPTTDIVLV 68
Query: 373 GPAKE----------------------EKKKDESSKPDQP-------------------- 320
GPAKE E KK+E +K ++P
Sbjct: 69 GPAKEPEKEKKEEPKKEEEKKEEPKKEEPKKEEEAKKEEPKKEGEKKEEPKKEEGKKEEE 128
Query: 319 KLPEPVKFYEAYPLYYQTRPSQYTQYHYVQSVEEDNVGCVIC 194
K P+PV E Y Q P Q T Y+YVQS+EE+ C IC
Sbjct: 129 KKPDPV--LELVKAYKQYNP-QMTTYYYVQSIEENPNACAIC 167
>pir||F86145 F22L4.5 protein - Arabidopsis thaliana
gi|8920587|gb|AAF81309.1|AC061957_5 Contains a weak
similarity to a farnesylated protein GMFP5 mRNA from
Glycine max gb|U64916. ESTs gb|AI993148, gb|T44360 come
from this gene. [Arabidopsis thaliana]
Length = 203
Score = 95.5 bits (236), Expect = 4e-19
Identities = 63/170 (37%), Positives = 82/170 (48%), Gaps = 51/170 (30%)
Frame = -2
Query: 550 ELHDDKIKQKAMKSVSXISGXESVSVDMGSKKLTLTGDIDPVHVVSKLRK-WCHTEIVSV 374
+LHDD+ KQKA+K+VS + G +S+++DM KKLT+ G +DPV+VVSKLRK W T+IV V
Sbjct: 35 DLHDDRAKQKALKTVSTLPGIDSIAMDMKEKKLTVIGTVDPVNVVSKLRKYWPMTDIVLV 94
Query: 373 GPAKEEKK--------------------------------------------KDESSKPD 326
GPAKE +K K E K D
Sbjct: 95 GPAKEPEKEKKEEPKKEGGGEPPKKEGEAPKEEGKKEGEAPKKEEEKKEGGDKKEGEKKD 154
Query: 325 QPK------LPEPVKFYEAYPLYYQTRPSQYTQYHYVQSVEEDNVGCVIC 194
QP+ +P P E Y P T Y+Y QS+EE+ CVIC
Sbjct: 155 QPQAQPQPVVPPPDHVLELVKAYKAYNP-HLTTYYYAQSIEENPNACVIC 203
>ref|NP_171656.1| expressed protein; protein id: At1g01490.1, supported by cDNA:
gi_17380661 [Arabidopsis thaliana]
gi|1922242|emb|CAA71173.1| hypothetical protein
[Arabidopsis thaliana] gi|17380662|gb|AAL36161.1|
unknown protein [Arabidopsis thaliana]
gi|20258999|gb|AAM14215.1| unknown protein [Arabidopsis
thaliana]
Length = 177
Score = 95.5 bits (236), Expect = 4e-19
Identities = 63/170 (37%), Positives = 82/170 (48%), Gaps = 51/170 (30%)
Frame = -2
Query: 550 ELHDDKIKQKAMKSVSXISGXESVSVDMGSKKLTLTGDIDPVHVVSKLRK-WCHTEIVSV 374
+LHDD+ KQKA+K+VS + G +S+++DM KKLT+ G +DPV+VVSKLRK W T+IV V
Sbjct: 9 DLHDDRAKQKALKTVSTLPGIDSIAMDMKEKKLTVIGTVDPVNVVSKLRKYWPMTDIVLV 68
Query: 373 GPAKEEKK--------------------------------------------KDESSKPD 326
GPAKE +K K E K D
Sbjct: 69 GPAKEPEKEKKEEPKKEGGGEPPKKEGEAPKEEGKKEGEAPKKEEEKKEGGDKKEGEKKD 128
Query: 325 QPK------LPEPVKFYEAYPLYYQTRPSQYTQYHYVQSVEEDNVGCVIC 194
QP+ +P P E Y P T Y+Y QS+EE+ CVIC
Sbjct: 129 QPQAQPQPVVPPPDHVLELVKAYKAYNP-HLTTYYYAQSIEENPNACVIC 177
>gb|AAM88621.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 179
Score = 77.8 bits (190), Expect = 8e-14
Identities = 39/69 (56%), Positives = 52/69 (74%)
Frame = -2
Query: 547 LHDDKIKQKAMKSVSXISGXESVSVDMGSKKLTLTGDIDPVHVVSKLRKWCHTEIVSVGP 368
+HD KQKAMK+VS + G + +S+DM S+K+T+ G +DPV+VVSKLRK I SVGP
Sbjct: 11 VHDKAEKQKAMKAVSALIGIDELSMDMASQKMTVIGMVDPVNVVSKLRKSWAATIESVGP 70
Query: 367 AKEEKKKDE 341
AKE +KK+E
Sbjct: 71 AKEPEKKEE 79
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 423,789,457
Number of Sequences: 1393205
Number of extensions: 8539806
Number of successful extensions: 25651
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 24313
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25573
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19234190289
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)