Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000976A_C02 KMC000976A_c02
(530 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
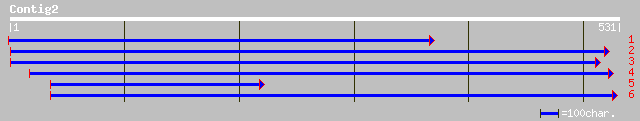
ref|NP_680210.1| similar to alanine--tRNA ligase, putative; prot... 132 3e-30
ref|NP_486458.1| alanyl-tRNA synthetase [Nostoc sp. PCC 7120] gi... 103 1e-21
ref|ZP_00072711.1| hypothetical protein [Trichodesmium erythraeu... 101 5e-21
gb|ZP_00107033.1| hypothetical protein [Nostoc punctiforme] 99 3e-20
ref|NP_441845.1| alanyl-tRNA synthetase [Synechocystis sp. PCC 6... 94 8e-19
>ref|NP_680210.1| similar to alanine--tRNA ligase, putative; protein id: At5g22800.1
[Arabidopsis thaliana] gi|10177227|dbj|BAB10601.1|
alanyl-tRNA synthetase [Arabidopsis thaliana]
Length = 954
Score = 132 bits (332), Expect = 3e-30
Identities = 65/80 (81%), Positives = 72/80 (89%)
Frame = -3
Query: 528 AVVLGSCPGEGKVSLVAAFTPGVVDRGIQAGKFIGQIAKLCGGGGGGRPNFAQAGGRKPE 349
AVVLGS P + KVSLVAAF+PGVV G+QAGKFIG IAKLCGGGGGG+PNFAQAGGRKPE
Sbjct: 873 AVVLGSSPEKDKVSLVAAFSPGVVSLGVQAGKFIGPIAKLCGGGGGGKPNFAQAGGRKPE 932
Query: 348 NLTSALEKAKSELIATLSEK 289
NL SALEKA+ +L+ATLSEK
Sbjct: 933 NLPSALEKAREDLVATLSEK 952
>ref|NP_486458.1| alanyl-tRNA synthetase [Nostoc sp. PCC 7120]
gi|20532251|sp|Q8YUD4|SYA_ANASP Alanyl-tRNA synthetase
(Alanine--tRNA ligase) (AlaRS) gi|25293093|pir||AC2108
alanyl-tRNA synthetase [imported] - Nostoc sp. (strain
PCC 7120) gi|17131510|dbj|BAB74117.1| alanyl-tRNA
synthetase [Nostoc sp. PCC 7120]
Length = 880
Score = 103 bits (258), Expect = 1e-21
Identities = 53/77 (68%), Positives = 61/77 (78%)
Frame = -3
Query: 528 AVVLGSCPGEGKVSLVAAFTPGVVDRGIQAGKFIGQIAKLCGGGGGGRPNFAQAGGRKPE 349
AVVLGS P KVSLVAAF+P V +G+QAGKFIG IAK+CGGGGGGRPN AQAGGR
Sbjct: 803 AVVLGSVPEADKVSLVAAFSPEVNKKGLQAGKFIGAIAKICGGGGGGRPNLAQAGGRDAS 862
Query: 348 NLTSALEKAKSELIATL 298
L +ALE+A+SEL + L
Sbjct: 863 KLPAALEQAQSELKSAL 879
>ref|ZP_00072711.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 897
Score = 101 bits (252), Expect = 5e-21
Identities = 52/74 (70%), Positives = 58/74 (78%)
Frame = -3
Query: 528 AVVLGSCPGEGKVSLVAAFTPGVVDRGIQAGKFIGQIAKLCGGGGGGRPNFAQAGGRKPE 349
AVVLGS EGKVSLVAAF+ V +G+QAGKFIG IAK+CGGGGGGRPN AQAGGR P
Sbjct: 821 AVVLGSA-AEGKVSLVAAFSKSVNGKGLQAGKFIGGIAKICGGGGGGRPNLAQAGGRDPS 879
Query: 348 NLTSALEKAKSELI 307
L ALE AK +L+
Sbjct: 880 KLKEALESAKEQLV 893
>gb|ZP_00107033.1| hypothetical protein [Nostoc punctiforme]
Length = 880
Score = 99.0 bits (245), Expect = 3e-20
Identities = 49/78 (62%), Positives = 60/78 (76%)
Frame = -3
Query: 528 AVVLGSCPGEGKVSLVAAFTPGVVDRGIQAGKFIGQIAKLCGGGGGGRPNFAQAGGRKPE 349
AVVLGS P KVS+VAAF+P V +G+QAGKF+G IAK+CGGGGGGRPN AQAGGR
Sbjct: 803 AVVLGSVPEADKVSIVAAFSPEVNKKGLQAGKFVGAIAKICGGGGGGRPNLAQAGGRDAS 862
Query: 348 NLTSALEKAKSELIATLS 295
L AL +A+S+L + L+
Sbjct: 863 KLPDALGQAESDLKSALA 880
>ref|NP_441845.1| alanyl-tRNA synthetase [Synechocystis sp. PCC 6803]
gi|2500959|sp|P74423|SYA_SYNY3 Alanyl-tRNA synthetase
(Alanine--tRNA ligase) (AlaRS) gi|7437614|pir||S76394
hypothetical protein - Synechocystis sp. (strain PCC
6803) gi|1653611|dbj|BAA18523.1| alanyl-tRNA synthetase
[Synechocystis sp. PCC 6803]
Length = 877
Score = 94.4 bits (233), Expect = 8e-19
Identities = 50/78 (64%), Positives = 58/78 (74%), Gaps = 1/78 (1%)
Frame = -3
Query: 528 AVVLGSCPGEGKVSLVAAFTPGVVD-RGIQAGKFIGQIAKLCGGGGGGRPNFAQAGGRKP 352
AVVL S P EGKVSLVAAF+P +V + ++AG+FIG IAK+CGGGGGGRPN AQAGGR
Sbjct: 799 AVVLASIPEEGKVSLVAAFSPQLVKTKQLKAGQFIGAIAKICGGGGGGRPNLAQAGGRDA 858
Query: 351 ENLTSALEKAKSELIATL 298
L AL AK L+A L
Sbjct: 859 SKLPEALATAKQTLLAEL 876
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 474,892,806
Number of Sequences: 1393205
Number of extensions: 10819044
Number of successful extensions: 52242
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 33744
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44643
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17596710992
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)