Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
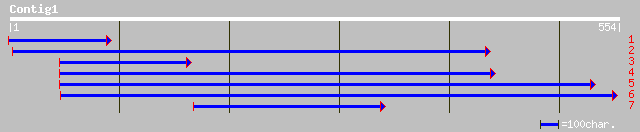
Query= KMC000957A_C01 KMC000957A_c01
(549 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||C96703 hypothetical protein T23K23.13 [imported] - Arabidop... 123 1e-27
gb|AAN52740.1| Putative trehalose-6-phosphate synthase [Oryza sa... 114 6e-25
ref|NP_567538.1| trehalose-6-phosphate synthase like protein; pr... 110 9e-24
pir||H71447 trehalose-6-phosphate synthase homolog DL4920W - Ara... 110 9e-24
ref|NP_176221.1| trehalose-6-phosphate synthase, putative; prote... 100 1e-20
>pir||C96703 hypothetical protein T23K23.13 [imported] - Arabidopsis thaliana
gi|12324075|gb|AAG52003.1|AC012563_13 putative
trehalose-6-phosphate synthase; 46897-44149 [Arabidopsis
thaliana]
Length = 847
Score = 123 bits (309), Expect = 1e-27
Identities = 58/75 (77%), Positives = 67/75 (89%)
Frame = -3
Query: 547 QEKGMSPDFVLCIGDDRSDEDMFEVITSSLAGPSIAPRAEVFACTVGRKPSKAKYYLDDN 368
QE+G P+FVLCIGDDRSDEDMFEVI SS GPSIAPRAE+FACTVG+KPSKAKYYLDD
Sbjct: 768 QERGTLPEFVLCIGDDRSDEDMFEVICSSTEGPSIAPRAEIFACTVGQKPSKAKYYLDDT 827
Query: 367 TDIVRMVPGLACVSE 323
T+IVR++ GLA V++
Sbjct: 828 TEIVRLMHGLASVTD 842
>gb|AAN52740.1| Putative trehalose-6-phosphate synthase [Oryza sativa (japonica
cultivar-group)]
Length = 860
Score = 114 bits (286), Expect = 6e-25
Identities = 54/74 (72%), Positives = 63/74 (84%)
Frame = -3
Query: 547 QEKGMSPDFVLCIGDDRSDEDMFEVITSSLAGPSIAPRAEVFACTVGRKPSKAKYYLDDN 368
QE+GM DFVLCIGDDRSDE+MF++ITSS G S+A AEVFACTVGRKPSKAKYYLDD
Sbjct: 772 QERGMCTDFVLCIGDDRSDEEMFQMITSSTCGESLAATAEVFACTVGRKPSKAKYYLDDT 831
Query: 367 TDIVRMVPGLACVS 326
++VR++ GLA VS
Sbjct: 832 AEVVRLMQGLASVS 845
>ref|NP_567538.1| trehalose-6-phosphate synthase like protein; protein id:
At4g17770.1, supported by cDNA: 95947. [Arabidopsis
thaliana] gi|26452424|dbj|BAC43297.1| putative
trehalose-6-phosphate synthase [Arabidopsis thaliana]
gi|29029046|gb|AAO64902.1| At4g17770 [Arabidopsis
thaliana]
Length = 862
Score = 110 bits (276), Expect = 9e-24
Identities = 50/74 (67%), Positives = 62/74 (83%)
Frame = -3
Query: 547 QEKGMSPDFVLCIGDDRSDEDMFEVITSSLAGPSIAPRAEVFACTVGRKPSKAKYYLDDN 368
QEKG DF+LC+GDDRSDEDMFEVI S+ GP+++P AE+FACTVG+KPSKAKYYLDD
Sbjct: 770 QEKGKLLDFILCVGDDRSDEDMFEVIMSAKDGPALSPVAEIFACTVGQKPSKAKYYLDDT 829
Query: 367 TDIVRMVPGLACVS 326
+I+RM+ GLA +
Sbjct: 830 AEIIRMLDGLAATN 843
>pir||H71447 trehalose-6-phosphate synthase homolog DL4920W - Arabidopsis
thaliana gi|2245136|emb|CAB10557.1|
trehalose-6-phosphate synthase like protein [Arabidopsis
thaliana] gi|7268530|emb|CAB78780.1|
trehalose-6-phosphate synthase like protein [Arabidopsis
thaliana]
Length = 865
Score = 110 bits (276), Expect = 9e-24
Identities = 50/74 (67%), Positives = 62/74 (83%)
Frame = -3
Query: 547 QEKGMSPDFVLCIGDDRSDEDMFEVITSSLAGPSIAPRAEVFACTVGRKPSKAKYYLDDN 368
QEKG DF+LC+GDDRSDEDMFEVI S+ GP+++P AE+FACTVG+KPSKAKYYLDD
Sbjct: 773 QEKGKLLDFILCVGDDRSDEDMFEVIMSAKDGPALSPVAEIFACTVGQKPSKAKYYLDDT 832
Query: 367 TDIVRMVPGLACVS 326
+I+RM+ GLA +
Sbjct: 833 AEIIRMLDGLAATN 846
>ref|NP_176221.1| trehalose-6-phosphate synthase, putative; protein id: At1g60140.1
[Arabidopsis thaliana] gi|7488402|pir||T02267
trehalose-6-phosphate synthase homolog T13D8.4 -
Arabidopsis thaliana gi|3249064|gb|AAC24048.1| Strong
similarity to trehalose-6-phosphate synthase homolog
gb|2245136 from A. thaliana chromosome 4 contig
gb|Z97344. [Arabidopsis thaliana]
gi|27462090|gb|AAO15312.1| trehalose-6-phosphate
synthase 3 [Arabidopsis thaliana]
Length = 861
Score = 100 bits (249), Expect = 1e-20
Identities = 45/75 (60%), Positives = 62/75 (82%)
Frame = -3
Query: 544 EKGMSPDFVLCIGDDRSDEDMFEVITSSLAGPSIAPRAEVFACTVGRKPSKAKYYLDDNT 365
E G++PDFV+CIGDDRSDE+MFE I+++L+ S + E+FACTVGRKPSKAKY+LD+ +
Sbjct: 771 EDGIAPDFVVCIGDDRSDEEMFENISTTLSAQSSSMSTEIFACTVGRKPSKAKYFLDEVS 830
Query: 364 DIVRMVPGLACVSEP 320
D+V+++ GLA S P
Sbjct: 831 DVVKLLQGLANTSSP 845
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 500,281,609
Number of Sequences: 1393205
Number of extensions: 11164814
Number of successful extensions: 28231
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 26719
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28205
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)