Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000954A_C01 KMC000954A_c01
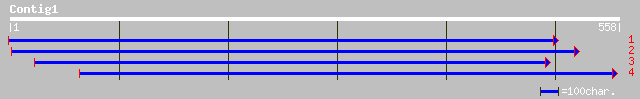
(558 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_188856.2| far-red impaired response protein, putative; pr... 79 4e-14
ref|NP_567455.1| Expressed protein; protein id: At4g15090.1, sup... 39 0.033
gb|AAD51282.1|AF159587_1 far-red impaired response protein [Arab... 39 0.033
pir||F71414 hypothetical protein - Arabidopsis thaliana gi|22448... 39 0.043
gb|EAA13897.1| agCP8235 [Anopheles gambiae str. PEST] 33 2.4
>ref|NP_188856.2| far-red impaired response protein, putative; protein id:
At3g22170.1 [Arabidopsis thaliana]
gi|11994736|dbj|BAB03065.1| far-red impaired response
protein; Mutator-like transposase-like protein;
phytochrome A signaling protein-like [Arabidopsis
thaliana]
Length = 839
Score = 79.0 bits (193), Expect = 4e-14
Identities = 40/67 (59%), Positives = 49/67 (72%), Gaps = 1/67 (1%)
Frame = -3
Query: 556 YGNQQTLQGLGPMSSIANSHDGYYGAHQNMPGLAQLDFLR-TSFTYGIRDDPNVRPGQLH 380
YGNQQT+QGL ++SIA S+D YYG Q + G +DF R +F+Y IRDDPNVR QLH
Sbjct: 774 YGNQQTMQGLRQLNSIAPSYDSYYGPQQGIHGQG-VDFFRPANFSYDIRDDPNVRTTQLH 832
Query: 379 EDPSRHA 359
ED SRH+
Sbjct: 833 EDASRHS 839
>ref|NP_567455.1| Expressed protein; protein id: At4g15090.1, supported by cDNA:
gi_19310413, supported by cDNA: gi_5764394 [Arabidopsis
thaliana]
Length = 768
Score = 39.3 bits (90), Expect = 0.033
Identities = 24/66 (36%), Positives = 41/66 (61%), Gaps = 2/66 (3%)
Frame = -3
Query: 556 YGNQQTLQGLGPMSSIANSHDGYYGAHQNMPGL-AQLDFL-RTSFTYGIRDDPNVRPGQL 383
Y +Q+T+QGLG ++SIA + D ++ Q M G+ Q+DF +FTY ++++ ++ QL
Sbjct: 702 YVDQRTIQGLGQLNSIAPAQDSFFTNQQAMSGMVGQIDFRPPPNFTYTLQEE-HLSSAQL 760
Query: 382 HEDPSR 365
SR
Sbjct: 761 PGSSSR 766
Score = 33.1 bits (74), Expect = 2.4
Identities = 16/34 (47%), Positives = 22/34 (64%)
Frame = -3
Query: 550 NQQTLQGLGPMSSIANSHDGYYGAHQNMPGLAQL 449
+QQ+LQ + +SS A +GYYG QN+ GL L
Sbjct: 660 SQQSLQPMETISSEAMDMNGYYGPQQNVQGLLNL 693
>gb|AAD51282.1|AF159587_1 far-red impaired response protein [Arabidopsis thaliana]
Length = 827
Score = 39.3 bits (90), Expect = 0.033
Identities = 24/66 (36%), Positives = 41/66 (61%), Gaps = 2/66 (3%)
Frame = -3
Query: 556 YGNQQTLQGLGPMSSIANSHDGYYGAHQNMPGL-AQLDFL-RTSFTYGIRDDPNVRPGQL 383
Y +Q+T+QGLG ++SIA + D ++ Q M G+ Q+DF +FTY ++++ ++ QL
Sbjct: 761 YVDQRTIQGLGQLNSIAPAQDSFFTNQQAMSGMVGQIDFRPPPNFTYTLQEE-HLSSAQL 819
Query: 382 HEDPSR 365
SR
Sbjct: 820 PGSSSR 825
Score = 33.1 bits (74), Expect = 2.4
Identities = 16/34 (47%), Positives = 22/34 (64%)
Frame = -3
Query: 550 NQQTLQGLGPMSSIANSHDGYYGAHQNMPGLAQL 449
+QQ+LQ + +SS A +GYYG QN+ GL L
Sbjct: 719 SQQSLQPMETISSEAMDMNGYYGPQQNVQGLLNL 752
>pir||F71414 hypothetical protein - Arabidopsis thaliana
gi|2244866|emb|CAB10288.1| hypothetical protein
[Arabidopsis thaliana] gi|7268255|emb|CAB78551.1|
hypothetical protein [Arabidopsis thaliana]
Length = 1705
Score = 38.9 bits (89), Expect = 0.043
Identities = 21/50 (42%), Positives = 32/50 (64%), Gaps = 2/50 (4%)
Frame = -3
Query: 556 YGNQQTLQGLGPMSSIANSHDGYYGAHQNMPGL-AQLDFL-RTSFTYGIR 413
YG QQ +QGLG ++SIA + D ++ Q M G+ Q+DF +FTY ++
Sbjct: 687 YGPQQNVQGLGQLNSIAPAQDSFFTNQQAMSGMVGQIDFRPPPNFTYTLQ 736
Score = 35.8 bits (81), Expect = 0.37
Identities = 17/35 (48%), Positives = 24/35 (68%)
Frame = -3
Query: 550 NQQTLQGLGPMSSIANSHDGYYGAHQNMPGLAQLD 446
+QQ+LQ + +SS A +GYYG QN+ GL QL+
Sbjct: 666 SQQSLQPMETISSEAMDMNGYYGPQQNVQGLGQLN 700
>gb|EAA13897.1| agCP8235 [Anopheles gambiae str. PEST]
Length = 3764
Score = 33.1 bits (74), Expect = 2.4
Identities = 15/39 (38%), Positives = 22/39 (55%)
Frame = +1
Query: 430 N*CAKNPAVPNQACSGVHHNNHHGCLLWNSLVPAPVESV 546
N C +NP PN AC+ V+H C ++ +VP+P V
Sbjct: 2901 NPCQENPCGPNAACTVVNH--RASCSCFSGMVPSPTAKV 2937
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 475,702,110
Number of Sequences: 1393205
Number of extensions: 10248013
Number of successful extensions: 20507
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 19872
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20498
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)