Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000952A_C04 KMC000952A_c04
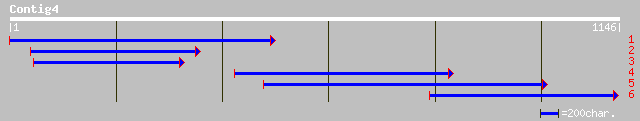
(1139 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||G86280 protein T5E21.13 [imported] - Arabidopsis thaliana g... 101 2e-20
gb|AAL91182.1| splicing factor, putative [Arabidopsis thaliana] 101 2e-20
ref|NP_172917.1| splicing factor, putative; protein id: At1g1465... 101 2e-20
ref|NP_568261.1| putative protein; protein id: At5g12280.1 [Arab... 79 1e-13
gb|EAA05190.1| ebiP8201 [Anopheles gambiae str. PEST] 55 3e-06
>pir||G86280 protein T5E21.13 [imported] - Arabidopsis thaliana
gi|7527720|gb|AAF63169.1|AC010657_5 T5E21.13 [Arabidopsis
thaliana]
Length = 1776
Score = 101 bits (252), Expect = 2e-20
Identities = 50/62 (80%), Positives = 55/62 (88%)
Frame = -2
Query: 1138 SLSETVARLKEKIAGDIQLPANQQKLSGKPGFLKDNLSLAHYHVSGGETLTLSLRERGGR 959
SLSE V LKEKIAG+IQ+PAN+QKLSGK GFLKDN+SLAHY+V GE LTLSLRERGGR
Sbjct: 1715 SLSENVGSLKEKIAGEIQIPANKQKLSGKAGFLKDNMSLAHYNVGAGEILTLSLRERGGR 1774
Query: 958 KR 953
KR
Sbjct: 1775 KR 1776
Score = 39.7 bits (91), Expect(2) = 4e-04
Identities = 18/25 (72%), Positives = 22/25 (88%)
Frame = -2
Query: 1138 SLSETVARLKEKIAGDIQLPANQQK 1064
SLSE V LKEKIAG++Q+PAN+QK
Sbjct: 858 SLSENVGSLKEKIAGEMQIPANKQK 882
Score = 26.9 bits (58), Expect(2) = 4e-04
Identities = 8/19 (42%), Positives = 14/19 (73%)
Frame = -1
Query: 1007 QRRRNTYPILKRAWW*KEM 951
+++RN Y + +R WW KE+
Sbjct: 880 KQKRNPYTVFERTWWEKEI 898
>gb|AAL91182.1| splicing factor, putative [Arabidopsis thaliana]
Length = 785
Score = 101 bits (252), Expect = 2e-20
Identities = 50/62 (80%), Positives = 55/62 (88%)
Frame = -2
Query: 1138 SLSETVARLKEKIAGDIQLPANQQKLSGKPGFLKDNLSLAHYHVSGGETLTLSLRERGGR 959
SLSE V LKEKIAG+IQ+PAN+QKLSGK GFLKDN+SLAHY+V GE LTLSLRERGGR
Sbjct: 724 SLSENVGSLKEKIAGEIQIPANKQKLSGKAGFLKDNMSLAHYNVGAGEILTLSLRERGGR 783
Query: 958 KR 953
KR
Sbjct: 784 KR 785
>ref|NP_172917.1| splicing factor, putative; protein id: At1g14650.1, supported by
cDNA: gi_19698892 [Arabidopsis thaliana]
Length = 785
Score = 101 bits (252), Expect = 2e-20
Identities = 50/62 (80%), Positives = 55/62 (88%)
Frame = -2
Query: 1138 SLSETVARLKEKIAGDIQLPANQQKLSGKPGFLKDNLSLAHYHVSGGETLTLSLRERGGR 959
SLSE V LKEKIAG+IQ+PAN+QKLSGK GFLKDN+SLAHY+V GE LTLSLRERGGR
Sbjct: 724 SLSENVGSLKEKIAGEIQIPANKQKLSGKAGFLKDNMSLAHYNVGAGEILTLSLRERGGR 783
Query: 958 KR 953
KR
Sbjct: 784 KR 785
>ref|NP_568261.1| putative protein; protein id: At5g12280.1 [Arabidopsis thaliana]
gi|14586358|emb|CAC42889.1| putative protein [Arabidopsis
thaliana]
Length = 419
Score = 79.0 bits (193), Expect = 1e-13
Identities = 39/53 (73%), Positives = 43/53 (80%)
Frame = -2
Query: 1138 SLSETVARLKEKIAGDIQLPANQQKLSGKPGFLKDNLSLAHYHVSGGETLTLS 980
SLSE VA LKEKI+G+IQ P N+QKL GK GFLKDN SLAHY+V GE LTLS
Sbjct: 365 SLSENVASLKEKISGEIQFPTNKQKLRGKAGFLKDNTSLAHYNVGAGEILTLS 417
>gb|EAA05190.1| ebiP8201 [Anopheles gambiae str. PEST]
Length = 675
Score = 54.7 bits (130), Expect = 3e-06
Identities = 25/62 (40%), Positives = 42/62 (67%)
Frame = -2
Query: 1138 SLSETVARLKEKIAGDIQLPANQQKLSGKPGFLKDNLSLAHYHVSGGETLTLSLRERGGR 959
+L++TV+ +K K+ + +P +QK+ F KDN ++AHY++ G T+ L L+ERGGR
Sbjct: 614 NLTDTVSSVKAKVQTETGMPPAKQKIFYDGMFFKDNNTIAHYNLLNGVTVALQLKERGGR 673
Query: 958 KR 953
K+
Sbjct: 674 KK 675
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 939,555,138
Number of Sequences: 1393205
Number of extensions: 20454002
Number of successful extensions: 49719
Number of sequences better than 10.0: 534
Number of HSP's better than 10.0 without gapping: 46170
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49514
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 69732809988
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)