Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000941A_C01 KMC000941A_c01
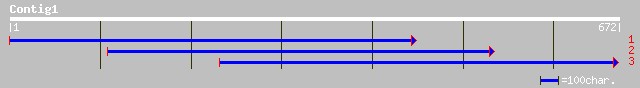
(672 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM28891.1| NADPH oxidase [Nicotiana tabacum] 33 2.7
gb|AAF73104.1|AF147783_1 whitefly-induced gp91-phox [Lycopersico... 33 2.7
emb|CAC84140.1| NADPH oxidase [Nicotiana tabacum] 33 2.7
dbj|BAC56865.1| respiratory burst oxidase homolog [Nicotiana ben... 33 2.7
gb|AAK71538.1| apolipoprotein H precursor [Pan troglodytes] 33 3.5
>gb|AAM28891.1| NADPH oxidase [Nicotiana tabacum]
Length = 938
Score = 33.5 bits (75), Expect = 2.7
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = -3
Query: 670 DFSHHTPPQFDFHKENF 620
DFSH T +FDFHKENF
Sbjct: 922 DFSHKTSTKFDFHKENF 938
>gb|AAF73104.1|AF147783_1 whitefly-induced gp91-phox [Lycopersicon esculentum]
gi|8131890|gb|AAF73124.1|AF148534_1 whitefly-induced
gp91-phox [Lycopersicon esculentum]
Length = 938
Score = 33.5 bits (75), Expect = 2.7
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = -3
Query: 670 DFSHHTPPQFDFHKENF 620
DFSH T +FDFHKENF
Sbjct: 922 DFSHKTSTKFDFHKENF 938
>emb|CAC84140.1| NADPH oxidase [Nicotiana tabacum]
Length = 939
Score = 33.5 bits (75), Expect = 2.7
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = -3
Query: 670 DFSHHTPPQFDFHKENF 620
DFSH T +FDFHKENF
Sbjct: 923 DFSHKTSTKFDFHKENF 939
>dbj|BAC56865.1| respiratory burst oxidase homolog [Nicotiana benthamiana]
Length = 939
Score = 33.5 bits (75), Expect = 2.7
Identities = 13/17 (76%), Positives = 14/17 (81%)
Frame = -3
Query: 670 DFSHHTPPQFDFHKENF 620
DFSH T +FDFHKENF
Sbjct: 923 DFSHKTSTKFDFHKENF 939
>gb|AAK71538.1| apolipoprotein H precursor [Pan troglodytes]
Length = 345
Score = 33.1 bits (74), Expect = 3.5
Identities = 13/35 (37%), Positives = 17/35 (48%)
Frame = -1
Query: 459 PEAKWFSCELATQSTRGGWTTTPACRHVMCKFKEK 355
P+ F + T +T G WT P CR V C F +
Sbjct: 176 PQHAMFGNDTITCTTHGNWTKLPECREVKCPFPSR 210
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 680,775,773
Number of Sequences: 1393205
Number of extensions: 17055505
Number of successful extensions: 35121
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 33801
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35110
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29421376608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)