Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
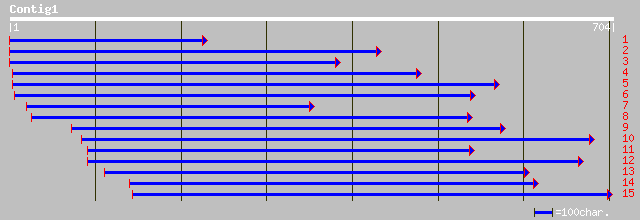
Query= KMC000936A_C01 KMC000936A_c01
(700 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_759468.1| Cell wall-associated hydrolase [Vibrio vulnific... 97 3e-32
ref|NP_799506.1| hypothetical protein [Bacillus megaterium] gi|2... 54 5e-19
pir||F81738 hypothetical protein TC0114 [imported] - Chlamydia m... 75 2e-18
ref|NP_296498.2| hypothetical protein [Chlamydia muridarum] gi|1... 74 3e-18
ref|NP_799507.1| hypothetical protein [Bacillus megaterium] gi|2... 93 3e-18
>ref|NP_759468.1| Cell wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27364363|ref|NP_759891.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27364369|ref|NP_759897.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27364408|ref|NP_759936.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27364496|ref|NP_760024.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27364598|ref|NP_760126.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27364804|ref|NP_760332.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27364878|ref|NP_760406.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27367804|ref|NP_763331.1| Cell wall-associated
hydrolase [Vibrio vulnificus CMCP6]
gi|27359377|gb|AAO08321.1|AE016813_73 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27360057|gb|AAO08995.1|AE016798_155 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27360482|gb|AAO09418.1|AE016800_23 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27360488|gb|AAO09424.1|AE016800_29 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27360527|gb|AAO09463.1|AE016800_68 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27360615|gb|AAO09551.1|AE016800_156 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27360717|gb|AAO09653.1|AE016800_258 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27360949|gb|AAO09859.1|AE016801_178 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
gi|27361023|gb|AAO09933.1|AE016801_252 Cell
wall-associated hydrolase [Vibrio vulnificus CMCP6]
Length = 144
Score = 97.4 bits (241), Expect(3) = 3e-32
Identities = 51/73 (69%), Positives = 54/73 (73%)
Frame = +1
Query: 343 PWDLLQPQDVMSRHRGAKRLRR*ELLGVISLLSPAYL*SVEREPFHTGLPDHYGRLSSLF 522
PWD LQPQDVMSRHRGAK RR ELLG ISLLSP YL SVER PFH+ PDHY LS L
Sbjct: 71 PWDRLQPQDVMSRHRGAKHRRRYELLGGISLLSPEYLLSVERWPFHSEPPDHYDLLSHLL 130
Query: 523 DQSVSQSGRLIPL 561
+ S SQ L+PL
Sbjct: 131 EPSFSQLSGLMPL 143
Score = 46.6 bits (109), Expect(3) = 3e-32
Identities = 22/27 (81%), Positives = 24/27 (88%)
Frame = +3
Query: 258 TPTVDRNRTVSRRSKPNSRTT*IGEQP 338
TPT DR+RTVSRRSKP+SRTT GEQP
Sbjct: 43 TPTADRDRTVSRRSKPSSRTTLNGEQP 69
Score = 37.4 bits (85), Expect(3) = 3e-32
Identities = 21/46 (45%), Positives = 26/46 (55%)
Frame = +2
Query: 134 AAVLPYQLSSPALLLA*QPVHHRLAQPSPLVLGLAPRSSPFNTNGR 271
+AV+ +LS A+ LA QP H R PLVLG AP + P T R
Sbjct: 3 SAVIDSELSYRAMRLATQPEHQRFVHSGPLVLGAAPFNLPTPTADR 48
>ref|NP_799506.1| hypothetical protein [Bacillus megaterium]
gi|28848595|gb|AAO52795.1| hypothetical protein
[Bacillus megaterium]
Length = 105
Score = 54.3 bits (129), Expect(3) = 5e-19
Identities = 27/34 (79%), Positives = 27/34 (79%)
Frame = +1
Query: 343 PWDLLQPQDVMSRHRGAKRLRR*ELLGVISLLSP 444
PWD LQPQD MSRHRGAK RR LLG ISLLSP
Sbjct: 71 PWDRLQPQDAMSRHRGAKPPRRCGLLGEISLLSP 104
Score = 44.7 bits (104), Expect(3) = 5e-19
Identities = 20/27 (74%), Positives = 24/27 (88%)
Frame = +3
Query: 258 TPTVDRNRTVSRRSKPNSRTT*IGEQP 338
TPT DR+RTVSRRS+P+SRT +GEQP
Sbjct: 43 TPTTDRDRTVSRRSEPSSRTALMGEQP 69
Score = 37.4 bits (85), Expect(3) = 5e-19
Identities = 21/46 (45%), Positives = 25/46 (53%)
Frame = +2
Query: 134 AAVLPYQLSSPALLLA*QPVHHRLAQPSPLVLGLAPRSSPFNTNGR 271
+A++P S PA+ LA Q VH R P PLVL AP P T R
Sbjct: 3 SALIPSTHSYPAMPLARQLVHQRCVHPGPLVLRTAPLKFPTPTTDR 48
>pir||F81738 hypothetical protein TC0114 [imported] - Chlamydia muridarum
(strain Nigg)
Length = 122
Score = 74.7 bits (182), Expect(2) = 2e-18
Identities = 38/52 (73%), Positives = 39/52 (74%)
Frame = +1
Query: 343 PWDLLQPQDVMSRHRGAKRLRR*ELLGVISLLSPAYL*SVEREPFHTGLPDH 498
PWDLLQPQD MSRHRGAK RR ELL ISLLSP YL SV+R FH PDH
Sbjct: 71 PWDLLQPQDAMSRHRGAKPPRRYELLVAISLLSPEYLLSVKRRRFHFPRPDH 122
Score = 39.7 bits (91), Expect(2) = 2e-18
Identities = 21/37 (56%), Positives = 26/37 (69%), Gaps = 3/37 (8%)
Frame = +3
Query: 237 LLAVLPL---TPTVDRNRTVSRRSKPNSRTT*IGEQP 338
+L PL PT DR++TVSRR +P+SRT IGEQP
Sbjct: 33 VLGTAPLKYPAPTKDRDQTVSRRFEPSSRTALIGEQP 69
Score = 38.5 bits (88), Expect = 0.091
Identities = 20/40 (50%), Positives = 22/40 (55%)
Frame = +2
Query: 152 QLSSPALLLA*QPVHHRLAQPSPLVLGLAPRSSPFNTNGR 271
+LS A+LLA QP HH PLVLG AP P T R
Sbjct: 9 ELSYSAMLLAKQPTHHWFVHSGPLVLGTAPLKYPAPTKDR 48
>ref|NP_296498.2| hypothetical protein [Chlamydia muridarum] gi|11282814|pir||G81737
hypothetical protein TC0130 [imported] - Chlamydia
muridarum (strain Nigg) gi|29251569|gb|AAF38993.2|
hypothetical protein [Chlamydia muridarum]
Length = 122
Score = 74.3 bits (181), Expect(2) = 3e-18
Identities = 38/52 (73%), Positives = 39/52 (74%)
Frame = +1
Query: 343 PWDLLQPQDVMSRHRGAKRLRR*ELLGVISLLSPAYL*SVEREPFHTGLPDH 498
PWDLLQPQD MSRHRGAK RR ELL ISLLSP YL SV+R FH PDH
Sbjct: 71 PWDLLQPQDAMSRHRGAKPPRRYELLVAISLLSPEYLLSVKRRRFHFPPPDH 122
Score = 39.7 bits (91), Expect(2) = 3e-18
Identities = 21/37 (56%), Positives = 26/37 (69%), Gaps = 3/37 (8%)
Frame = +3
Query: 237 LLAVLPL---TPTVDRNRTVSRRSKPNSRTT*IGEQP 338
+L PL PT DR++TVSRR +P+SRT IGEQP
Sbjct: 33 VLGTAPLKYPAPTKDRDQTVSRRFEPSSRTALIGEQP 69
Score = 38.5 bits (88), Expect = 0.091
Identities = 20/40 (50%), Positives = 22/40 (55%)
Frame = +2
Query: 152 QLSSPALLLA*QPVHHRLAQPSPLVLGLAPRSSPFNTNGR 271
+LS A+LLA QP HH PLVLG AP P T R
Sbjct: 9 ELSYSAMLLAKQPTHHWFVHSGPLVLGTAPLKYPAPTKDR 48
>ref|NP_799507.1| hypothetical protein [Bacillus megaterium]
gi|28848605|gb|AAO52805.1| hypothetical protein
[Bacillus megaterium]
Length = 95
Score = 93.2 bits (230), Expect = 3e-18
Identities = 51/89 (57%), Positives = 56/89 (62%), Gaps = 8/89 (8%)
Frame = -1
Query: 544 LTVRPTGRTETKVGHSDPGVPCGRALAQRIKGTPGITG**LPRALIDGVVWHLDVDSSHP 365
+T R T R TKVG SDP VP GRA+AQRIK TPGITG PR IDG VWHLDV SSHP
Sbjct: 1 MTARTTSRAGTKVGLSDPVVPHGRAIAQRIKATPGITGLSPPRVHIDGEVWHLDVGSSHP 60
Query: 364 GVEEGPKALGCSPIQ--------VVRELG 302
G GPK P++ VVR+ G
Sbjct: 61 GAVVGPKGWAVRPLKRYASWVQNVVRQFG 89
Score = 33.5 bits (75), Expect = 2.9
Identities = 16/26 (61%), Positives = 17/26 (64%)
Frame = -2
Query: 348 PRLSVVRRFKWYVSWV*NVVRQFGSY 271
P+ VR K Y SWV NVVRQFG Y
Sbjct: 66 PKGWAVRPLKRYASWVQNVVRQFGPY 91
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 663,353,189
Number of Sequences: 1393205
Number of extensions: 15822616
Number of successful extensions: 52270
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 44164
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 51548
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31967457276
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)