Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC000927A_C01 KMC000927A_c01
(558 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
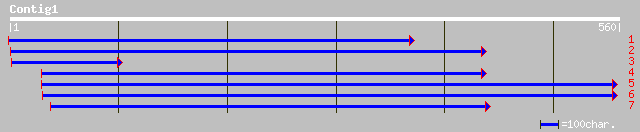
emb|CAD20127.2| raffinose synthase [Pisum sativum] 123 1e-27
gb|AAD02832.1| raffinose synthase [Cucumis sativus] 120 1e-26
ref|NP_198855.1| glycosyl hydrolase family 36; protein id: At5g4... 113 2e-24
dbj|BAB64768.1| putative raffinose synthase [Oryza sativa (japon... 89 4e-17
gb|AAL90901.1| AT5g20250/F5O24_140 [Arabidopsis thaliana] gi|233... 66 3e-10
>emb|CAD20127.2| raffinose synthase [Pisum sativum]
Length = 798
Score = 123 bits (309), Expect = 1e-27
Identities = 57/104 (54%), Positives = 80/104 (76%)
Frame = -2
Query: 554 FELVTVSPVVTASLASKSVQFAPVGLVNMLNTGGAIPSLDFDEAQNLVPVGVRGTGEMRV 375
FEL+TVSP+ S + +QFAP+GLVNMLN+GGA+ SL+FD++ +LV +GVRG GE+ V
Sbjct: 697 FELMTVSPLKVFS--KRLIQFAPIGLVNMLNSGGAVQSLEFDDSASLVKIGVRGCGELSV 754
Query: 374 FSSERPSACRIDGKEADFEYEGSMVIIQVPWPSSSKLSYVQYIF 243
F+SE+P C+IDG +F+YE MV +Q+ WP SS LS V+++F
Sbjct: 755 FASEKPVCCKIDGVSVEFDYEDKMVRVQILWPGSSTLSLVEFLF 798
>gb|AAD02832.1| raffinose synthase [Cucumis sativus]
Length = 784
Score = 120 bits (301), Expect = 1e-26
Identities = 59/109 (54%), Positives = 82/109 (75%), Gaps = 4/109 (3%)
Frame = -2
Query: 557 DFELVTVSPVVTASLASKSVQFAPVGLVNMLNTGGAIPSLDFDEAQNLVPVGVRGTGEMR 378
+FEL+TVSPV L S+ FAP+GLVNMLNT GAI S+D+D+ + V +GV+G GEMR
Sbjct: 678 EFELITVSPVT--KLIQTSLHFAPIGLVNMLNTSGAIQSVDYDDDLSSVEIGVKGCGEMR 735
Query: 377 VFSSERPSACRIDGKEADFEY-EGSMVIIQVPWP---SSSKLSYVQYIF 243
VF+S++P ACRIDG++ F+Y + MV++QVPWP SS +S ++Y+F
Sbjct: 736 VFASKKPRACRIDGEDVGFKYDQDQMVVVQVPWPIDSSSGGISVIEYLF 784
>ref|NP_198855.1| glycosyl hydrolase family 36; protein id: At5g40390.1, supported by
cDNA: gi_17065409, supported by cDNA: gi_20148632
[Arabidopsis thaliana] gi|10178150|dbj|BAB11595.1|
raffinose synthase protein [Arabidopsis thaliana]
gi|17065410|gb|AAL32859.1| raffinose synthase protein
[Arabidopsis thaliana] gi|20148633|gb|AAM10207.1|
raffinose synthase protein [Arabidopsis thaliana]
Length = 783
Score = 113 bits (282), Expect = 2e-24
Identities = 58/104 (55%), Positives = 76/104 (72%)
Frame = -2
Query: 554 FELVTVSPVVTASLASKSVQFAPVGLVNMLNTGGAIPSLDFDEAQNLVPVGVRGTGEMRV 375
FEL+TVSPVVT + SV+FAP+GLVNMLNT GAI SL +++ V VGV G GE RV
Sbjct: 684 FELITVSPVVT--IEGNSVRFAPIGLVNMLNTSGAIRSLVYNDES--VEVGVFGAGEFRV 739
Query: 374 FSSERPSACRIDGKEADFEYEGSMVIIQVPWPSSSKLSYVQYIF 243
++S++P +C IDG+ +F YE SMV++QVPW LS +QY+F
Sbjct: 740 YASKKPVSCLIDGEVVEFGYEDSMVMVQVPWSGPDGLSSIQYLF 783
>dbj|BAB64768.1| putative raffinose synthase [Oryza sativa (japonica cultivar-group)]
gi|20804839|dbj|BAB92522.1| putative raffinose synthase
[Oryza sativa (japonica cultivar-group)]
Length = 816
Score = 89.0 bits (219), Expect = 4e-17
Identities = 45/107 (42%), Positives = 65/107 (60%), Gaps = 3/107 (2%)
Frame = -2
Query: 554 FELVTVSPVVTASLASKSVQFAPVGLVNMLNTGGAIPSLDFDEAQNLVP--VGVRGTGEM 381
+EL+ V+PV + FAP+GL NMLN GGA+ + V V V+G GEM
Sbjct: 710 YELLVVAPVRAIVSPELGIGFAPIGLANMLNAGGAVQGFEAARKDGDVAAEVAVKGAGEM 769
Query: 380 RVFSSERPSACRIDGKEADFEYEGSMVIIQVPWP-SSSKLSYVQYIF 243
+SS RP C+++G++A+F+YE +V + VPW SS KLS V+Y +
Sbjct: 770 VAYSSARPRLCKVNGQDAEFKYEDGIVTVDVPWTGSSKKLSRVEYFY 816
>gb|AAL90901.1| AT5g20250/F5O24_140 [Arabidopsis thaliana]
gi|23308457|gb|AAN18198.1| At5g20250/F5O24_140
[Arabidopsis thaliana]
Length = 844
Score = 65.9 bits (159), Expect = 3e-10
Identities = 34/88 (38%), Positives = 52/88 (58%)
Frame = -2
Query: 557 DFELVTVSPVVTASLASKSVQFAPVGLVNMLNTGGAIPSLDFDEAQNLVPVGVRGTGEMR 378
+ E+ TVSP+ S V FAP+GLVNM N+GGAI L ++ + V + V+G G+
Sbjct: 739 EHEIFTVSPI---SHLVDGVSFAPIGLVNMYNSGGAIEGLRYEAEKMKVVMEVKGCGKFG 795
Query: 377 VFSSERPSACRIDGKEADFEYEGSMVII 294
+SS +P C ++ E FEY+ S ++
Sbjct: 796 SYSSVKPKRCVVESNEIAFEYDSSSGLV 823
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 490,930,809
Number of Sequences: 1393205
Number of extensions: 10896666
Number of successful extensions: 38774
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 37014
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38717
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)